BETAWRAP: successful prediction of parallel beta -helices from primary sequence reveals an association with many microbial pathogens
- PMID: 11752429
- PMCID: PMC64942
- DOI: 10.1073/pnas.251267298
BETAWRAP: successful prediction of parallel beta -helices from primary sequence reveals an association with many microbial pathogens
Abstract
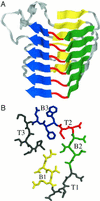
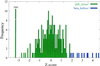
The amino acid sequence rules that specify beta-sheet structure in proteins remain obscure. A subclass of beta-sheet proteins, parallel beta-helices, represent a processive folding of the chain into an elongated topologically simpler fold than globular beta-sheets. In this paper, we present a computational approach that predicts the right-handed parallel beta-helix supersecondary structural motif in primary amino acid sequences by using beta-strand interactions learned from non-beta-helix structures. A program called BETAWRAP (http://theory.lcs.mit.edu/betawrap) implements this method and recognizes each of the seven known parallel beta-helix families, when trained on the known parallel beta-helices from outside that family. BETAWRAP identifies 2,448 sequences among 595,890 screened from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) nonredundant protein database as likely parallel beta-helices. It identifies surprisingly many bacterial and fungal protein sequences that play a role in human infectious disease; these include toxins, virulence factors, adhesins, and surface proteins of Chlamydia, Helicobacteria, Bordetella, Leishmania, Borrelia, Rickettsia, Neisseria, and Bacillus anthracis. Also unexpected was the rarity of the parallel beta-helix fold and its predicted sequences among higher eukaryotes. The computational method introduced here can be called a three-dimensional dynamic profile method because it generates interstrand pairwise correlations from a processive sequence wrap. Such methods may be applicable to recognizing other beta structures for which strand topology and profiles of residue accessibility are well conserved.
Figures
References
-
- Yoder M D, Keen N T, Jurnak F. Science. 1993;260:1503–1507. - PubMed
-
- Yoder M D, Lietzke S E, Jurnak F. Structure. 1993;1:241–251. - PubMed
-
- Jenkins J, Mayans O, Pickersgill R. J Struct Biol. 1998;122:236–246. - PubMed
-
- Murzin A G, Brenner S F, Hubbard T, Chothia C. J Mol Biol. 1995;297:536–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

