The free energy landscape for beta hairpin folding in explicit water
- PMID: 11752441
- PMCID: PMC64961
- DOI: 10.1073/pnas.201543998
The free energy landscape for beta hairpin folding in explicit water
Abstract
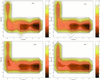
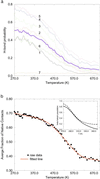
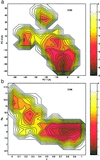
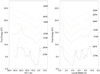
The folding free energy landscape of the C-terminal beta hairpin of protein G has been explored in this study with explicit solvent under periodic boundary condition and OPLSAA force field. A highly parallel replica exchange method that combines molecular dynamics trajectories with a temperature exchange Monte Carlo process is used for sampling with the help of a new efficient algorithm P3ME/RESPA. The simulation results show that the hydrophobic core and the beta strand hydrogen bond form at roughly the same time. The free energy landscape with respect to various reaction coordinates is found to be rugged at low temperatures and becomes a smooth funnel-like landscape at about 360 K. In contrast to some very recent studies, no significant helical content has been found in our simulation at all temperatures studied. The beta hairpin population and hydrogen-bond probability are in reasonable agreement with the experiment at biological temperature, but both decay more slowly than the experiment with temperature.
Figures

Similar articles
-
Trp-cage: folding free energy landscape in explicit water.Proc Natl Acad Sci U S A. 2003 Nov 11;100(23):13280-5. doi: 10.1073/pnas.2233312100. Epub 2003 Oct 27. Proc Natl Acad Sci U S A. 2003. PMID: 14581616 Free PMC article.
-
Can a continuum solvent model reproduce the free energy landscape of a beta -hairpin folding in water?Proc Natl Acad Sci U S A. 2002 Oct 1;99(20):12777-82. doi: 10.1073/pnas.142430099. Epub 2002 Sep 19. Proc Natl Acad Sci U S A. 2002. PMID: 12242327 Free PMC article.
-
Exploring the protein folding free energy landscape: coupling replica exchange method with P3ME/RESPA algorithm.J Mol Graph Model. 2004 May;22(5):451-63. doi: 10.1016/j.jmgm.2003.12.011. J Mol Graph Model. 2004. PMID: 15099840
-
Free energy landscape of protein folding in water: explicit vs. implicit solvent.Proteins. 2003 Nov 1;53(2):148-61. doi: 10.1002/prot.10483. Proteins. 2003. PMID: 14517967
-
Free energy landscape and folding mechanism of a beta-hairpin in explicit water: a replica exchange molecular dynamics study.Proteins. 2005 Dec 1;61(4):795-808. doi: 10.1002/prot.20696. Proteins. 2005. PMID: 16240446
Cited by
-
Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease.J Biol Chem. 2013 Jun 14;288(24):17643-53. doi: 10.1074/jbc.M113.452714. Epub 2013 Apr 26. J Biol Chem. 2013. PMID: 23625918 Free PMC article.
-
Nascent β-Hairpin Formation of a Natively Unfolded Peptide Reveals the Role of Hydrophobic Contacts.Biophys J. 2015 Aug 4;109(3):630-8. doi: 10.1016/j.bpj.2015.06.035. Biophys J. 2015. PMID: 26244744 Free PMC article.
-
Force-induced unzipping transitions in an athermal crowded environment.J Phys Chem B. 2013 Oct 24;117(42):13107-14. doi: 10.1021/jp402922q. Epub 2013 Jul 12. J Phys Chem B. 2013. PMID: 23789729 Free PMC article.
-
Synthesis, modeling, and pharmacological evaluation of UMB 425, a mixed μ agonist/δ antagonist opioid analgesic with reduced tolerance liabilities.ACS Chem Neurosci. 2013 Sep 18;4(9):1256-66. doi: 10.1021/cn4000428. Epub 2013 Jun 11. ACS Chem Neurosci. 2013. PMID: 23713721 Free PMC article.
-
Role of hydrophobic interactions and salt-bridges in beta-hairpin folding.J Mol Model. 2006 Jan;12(2):197-204. doi: 10.1007/s00894-005-0018-6. Epub 2005 Oct 18. J Mol Model. 2006. PMID: 16231193
References
-
- Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999.
-
- Blanco F J, Rivas G, Serrano L. Nat Struct Biol. 1994;1:584–590. - PubMed
-
- Blanco F J, Serrano L. Eur J Biochem. 1995;230:634–649. - PubMed
-
- Munoz V, Thompson P A, Hofrichter J, Eaton W A. Nature (London) 1997;390:196–199. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

