A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim
- PMID: 11752475
- PMCID: PMC65037
- DOI: 10.1073/pnas.261459698
A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim
Abstract
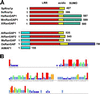
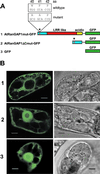
Ran is a small signaling GTPase that is involved in nucleocytoplasmic transport. Two additional functions of animal Ran in the formation of spindle asters and the reassembly of the nuclear envelope in mitotic cells have been recently reported. In contrast to Ras or Rho, Ran is not associated with membranes. Instead, the spatial sequestering of its accessory proteins, the Ran GTPase-activating protein RanGAP and the nucleotide exchange factor RCC1, appears to define the local concentration of RanGTP vs. RanGDP involved in signaling. Mammalian RanGAP is bound to the nuclear pore by a mechanism involving the attachment of small ubiquitin-related modifier protein (SUMO) to its C terminus and the subsequent binding of the SUMOylated domain to the nucleoporin Nup358. Here we show that plant RanGAP utilizes a different mechanism for nuclear envelope association, involving a novel targeting domain that appears to be unique to plants. The N-terminal WPP domain is highly conserved among plant RanGAPs and the small, plant-specific nuclear envelope-associated protein MAF1, but not present in yeast or animal RanGAP. Confocal laser scanning microscopy of green fluorescent protein (GFP) fusion proteins showed that it is necessary for RanGAP targeting and sufficient to target the heterologous protein GFP to the plant nuclear rim. The highly conserved tryptophan and proline residues of the WPP motif are necessary for its function. The 110-aa WPP domain is the first nuclear-envelope targeting domain identified in plants. Its fundamental difference to its mammalian counterpart implies that different mechanisms have evolved in plants and animals to anchor RanGAP at the nuclear surface.
Figures



Similar articles
-
Targeting proteins to the plant nuclear envelope.Biochem Soc Trans. 2010 Jun;38(3):733-40. doi: 10.1042/BST0380733. Biochem Soc Trans. 2010. PMID: 20491658 Review.
-
Plant-specific mitotic targeting of RanGAP requires a functional WPP domain.Plant J. 2005 Apr;42(2):270-82. doi: 10.1111/j.1365-313X.2005.02368.x. Plant J. 2005. PMID: 15807788
-
Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.Nature. 1999 Mar 4;398(6722):39-46. doi: 10.1038/17969. Nature. 1999. PMID: 10078529
-
Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP.Plant Cell. 2008 Jun;20(6):1639-51. doi: 10.1105/tpc.108.059220. Epub 2008 Jun 30. Plant Cell. 2008. PMID: 18591351 Free PMC article.
-
Going green: plants' alternative way to position the Ran gradient.J Microsc. 2008 Aug;231(2):225-33. doi: 10.1111/j.1365-2818.2008.02038.x. J Microsc. 2008. PMID: 18778420 Review.
Cited by
-
Color recovery after photoconversion of H2B::mEosFP allows detection of increased nuclear DNA content in developing plant cells.Plant Physiol. 2012 Jan;158(1):95-106. doi: 10.1104/pp.111.187062. Epub 2011 Nov 22. Plant Physiol. 2012. PMID: 22108524 Free PMC article.
-
The plant nuclear envelope.Planta. 2004 Jan;218(3):327-36. doi: 10.1007/s00425-003-1132-2. Epub 2003 Nov 11. Planta. 2004. PMID: 14610677 Review.
-
Protein interactions at the higher plant nuclear envelope: evidence for a linker of nucleoskeleton and cytoskeleton complex.Front Plant Sci. 2014 May 7;5:183. doi: 10.3389/fpls.2014.00183. eCollection 2014. Front Plant Sci. 2014. PMID: 24847341 Free PMC article. Review.
-
Preprophase band formation and cortical division zone establishment: RanGAP behaves differently from microtubules during their band formation.Plant Signal Behav. 2015;10(9):e1060385. doi: 10.1080/15592324.2015.1060385. Plant Signal Behav. 2015. PMID: 26237087 Free PMC article.
-
Proteomic approach to identify the differentially abundant proteins during flavour development in tuberous roots of Decalepis hamiltonii Wight & Arn.3 Biotech. 2021 Apr;11(4):173. doi: 10.1007/s13205-021-02714-x. Epub 2021 Mar 18. 3 Biotech. 2021. PMID: 33927964 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

