Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing
- PMID: 11756546
- PMCID: PMC139751
- DOI: 10.1128/MCB.22.2.505-516.2002
Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing
Abstract
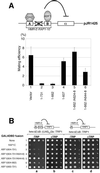
Autonomously replicating sequence-binding factor 1 (ABF1) is a multifunctional, site-specific DNA binding protein that is essential for cell viability in Saccharomyces cerevisiae. ABF1 plays a direct role in transcriptional activation, stimulation of DNA replication, and gene silencing at the mating-type loci. Here we demonstrate that all three activities of ABF1 are conferred by the C terminus of the protein (amino acids [aa] 604 to 731). Furthermore, a detailed mutational analysis has revealed two important clusters of amino acid residues in the C terminus (C-terminal sequence 1 [CS1], aa 624 to 628; and CS2, aa 639 to 662). While both regions play a pivotal role in supporting cell viability, they make distinct contributions to ABF1 functions in various nuclear processes. CS1 specifically participates in transcriptional silencing and/or repression in a context-dependent manner, whereas CS2 is universally required for all three functions of ABF1. When tethered to specific regions of the genome, a 30-aa fragment that contains CS2 alone is sufficient for activation of transcription and chromosomal replication. In addition, CS2 is responsible for ABF1-mediated chromatin remodeling. Based on these results, we suggest that ABF1 may function as a chromatin-reorganizing factor to increase accessibility of the local chromatin structure, which in turn facilitates the action of additional factors to establish either an active or repressed chromatin state.
Figures







Similar articles
-
Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae.Mol Cell Biol. 2004 Oct;24(20):9152-64. doi: 10.1128/MCB.24.20.9152-9164.2004. Mol Cell Biol. 2004. PMID: 15456886 Free PMC article.
-
Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae.Mol Cell Biol. 1992 Mar;12(3):1064-77. doi: 10.1128/mcb.12.3.1064-1077.1992. Mol Cell Biol. 1992. PMID: 1545789 Free PMC article.
-
Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene.Mol Cell Biol. 1996 Jun;16(6):2777-86. doi: 10.1128/MCB.16.6.2777. Mol Cell Biol. 1996. PMID: 8649386 Free PMC article.
-
Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae.J Biol Chem. 2004 Aug 13;279(33):34865-72. doi: 10.1074/jbc.M405156200. Epub 2004 Jun 10. J Biol Chem. 2004. PMID: 15192094
-
The BAH domain, polybromo and the RSC chromatin remodelling complex.Gene. 2001 May 2;268(1-2):1-7. doi: 10.1016/s0378-1119(01)00428-0. Gene. 2001. PMID: 11368894 Review.
Cited by
-
RNA Polymerase I and Fob1 contributions to transcriptional silencing at the yeast rDNA locus.Nucleic Acids Res. 2016 Jul 27;44(13):6173-84. doi: 10.1093/nar/gkw212. Epub 2016 Apr 8. Nucleic Acids Res. 2016. PMID: 27060141 Free PMC article.
-
Chromatin mediation of a transcriptional memory effect in yeast.G3 (Bethesda). 2015 Mar 5;5(5):829-38. doi: 10.1534/g3.115.017418. G3 (Bethesda). 2015. PMID: 25748434 Free PMC article.
-
Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae.Mol Cell Biol. 2004 Oct;24(20):9152-64. doi: 10.1128/MCB.24.20.9152-9164.2004. Mol Cell Biol. 2004. PMID: 15456886 Free PMC article.
-
Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae.Nucleic Acids Res. 2007;35(1):193-202. doi: 10.1093/nar/gkl1059. Epub 2006 Dec 7. Nucleic Acids Res. 2007. PMID: 17158163 Free PMC article.
-
Inference of transcription modification in long-live yeast strains from their expression profiles.BMC Genomics. 2007 Jul 6;8:219. doi: 10.1186/1471-2164-8-219. BMC Genomics. 2007. PMID: 17617911 Free PMC article.
References
-
- Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463–471 - PubMed
-
- Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15–19 - PubMed
-
- Buck, S. W., and D. Shore. 1995. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9:370–384 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
