Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer
- PMID: 11756685
- PMCID: PMC117556
- DOI: 10.1073/pnas.012610299
Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer
Abstract
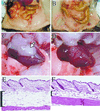
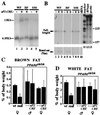
Targeting of the nuclear prostaglandin receptor peroxisome proliferator-activated receptor delta (PPARdelta) by homologous recombination results in placental defects and frequent (>90%) midgestation lethality. Surviving PPARdelta(-/-) mice exhibit a striking reduction in adiposity relative to wild-type levels. This effect is not reproduced in mice harboring an adipose tissue-specific deletion of PPARdelta, and thus likely reflects peripheral PPARdelta functions in systemic lipid metabolism. Finally, we observe that PPARdelta is dispensable for polyp formation in the intestine and colon of APC(min) mice, inconsistent with its recently proposed role in the establishment of colorectal tumors. Together, these observations reveal specific roles for PPARdelta in embryo development and adipocyte physiology, but not cancer.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

