Exploring the folding landscape of a structured RNA
- PMID: 11756689
- PMCID: PMC117531
- DOI: 10.1073/pnas.221593598
Exploring the folding landscape of a structured RNA
Abstract
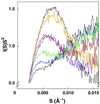
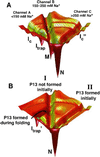
Structured RNAs achieve their active states by traversing complex, multidimensional energetic landscapes. Here we probe the folding landscape of the Tetrahymena ribozyme by using a powerful approach: the folding of single ribozyme molecules is followed beginning from distinct regions of the folding landscape. The experiments, combined with small-angle x-ray scattering results, show that the landscape contains discrete folding pathways. These pathways are separated by large free-energy barriers that prevent interconversion between them, indicating that the pathways lie in deep channels in the folding landscape. Chemical protection and mutagenesis experiments are then used to elucidate the structural features that determine which folding pathway is followed. Strikingly, a specific long-range tertiary contact can either help folding or hinder folding, depending on when it is formed during the process. Together these results provide an unprecedented view of the topology of an RNA folding landscape and the RNA structural features that underlie this multidimensional landscape.
Figures
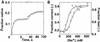



References
-
- Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. - PubMed
-
- Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. - PubMed
-
- Pan J, Woodson S A. J Mol Biol. 1998;280:597–609. - PubMed
-
- Treiber D K, Rook M S, Zarrinkar P P, Williamson J R. Science. 1998;279:1943–1946. - PubMed
-
- Rook M S, Treiber D K, Williamson J R. J Mol Biol. 1998;281:609–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

