Modeling of 5' nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica
- PMID: 11773092
- PMCID: PMC120126
- DOI: 10.1128/JCM.40.1.52-60.2002
Modeling of 5' nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica
Abstract
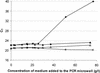
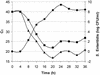
The performance of a 5' nuclease real-time PCR assay was studied to optimize an automated method of detection of preenriched Salmonella enterica cells in buffered peptone water (BPW). The concentrations and interactions of the PCR reagents were evaluated on the basis of two detection responses, the threshold cycle (C(T)) and the fluorescence intensity by a normalized reporter value (DeltaR(n)). The C(T) response was identified as the most suitable for detection modeling to describe the PCR performances of different samples. DNA extracted from S. enterica serovar Enteritidis was studied in double-distilled H2O (ddH2O) and in two different enrichment media (brain heart infusion and BPW) with two PCR mixtures based on AmpliTaq Gold or rTTH: A descriptive model was proposed and fitted to the available experimental data. Equivalent PCR performances for the two PCR mixtures were obtained when DNA was diluted in ddH2O. However, the level of detection of DNA was affected when BPW was present during amplification. Use of the rTth mixture generated a 1-log-unit wider linear range of amplification, and the DNA detection levels were 2 x 10(-13) g/microwell for the rTth mixture and 2 x 10(-12) g/microwell for the AmpliTaq Gold mixture. To verify the improved amplification capacity of the rTth mixture, BPW was inoculated with 1 CFU of S. enterica serovar Enteritidis per ml and the mixture was incubated at 30 degrees C. Samples for PCR were withdrawn every 4 h during a 36-h enrichment. Use of the rTth mixture resulted in an earlier PCR detection during enrichment than use of the AmpliTaq Gold mixture. For accurate detection (C(T) < or = 30) of S. enterica serovar Enteritidis inoculated in BPW, the rTth mixture required 8.4 h of enrichment, while the AmpliTaq Gold mixture needed 11.6 h. In conclusion, the principle applied can improve the methodology of 5' nuclease real-time PCR for numerical optimization of sample pretreatment strategies to provide automated diagnostic PCR procedures.
Figures



References
-
- Agresti, A. 1996. An introduction to categorical data analysis. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

