Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism
- PMID: 11773100
- PMCID: PMC120100
- DOI: 10.1128/JCM.40.1.101-104.2002
Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism
Abstract
Two molecular fingerprinting techniques, pulsed-field gel electrophoresis (PFGE) and amplified fragment length polymorphism (AFLP), were used to investigate the epidemiological relatedness among Clostridium difficile isolates from suspected outbreaks in three general hospitals. Analysis by PFGE yielded inconclusive data as a result of extensive DNA degradation. Although this degradation could be prevented to a certain extent by the inclusion of thiourea in the electrophoresis buffer, the weak DNA banding patterns obtained in this way were still far from optimal. AFLP data were obtained by using fluorescently labeled PCR primers and analysis on an ABI PRISM automated DNA analysis platform. AFLP analysis yielded high resolution and highly reproducible DNA fingerprinting patterns from which the epidemiological relatedness among the isolates could easily be determined. AFLP results could be readily obtained within 24 h, whereas 3 to 4 days were routinely required to complete the lengthy PFGE protocol. AFLP clearly proved to be a much more fail-safe fingerprinting method for C. difficile isolates, especially for those isolates for which a standard PFGE procedure yielded inconclusive results due to DNA degradation.
Figures

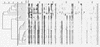
Comment in
-
Pulsed-field gel electrophoresis can yield DNA fingerprints of degradation-susceptible Clostridium difficile strains.J Clin Microbiol. 2002 Sep;40(9):3546-7; author reply 3547. doi: 10.1128/JCM.40.9.3546-3547.2002. J Clin Microbiol. 2002. PMID: 12202619 Free PMC article. No abstract available.
References
-
- Aarts, H. J., L. A. van Lith, and J. Keijer. 1998. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett. Appl. Microbiol. 26:131–135. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

