Phylogeny-based rapid identification of mycoplasmas and ureaplasmas from urethritis patients
- PMID: 11773101
- PMCID: PMC120092
- DOI: 10.1128/JCM.40.1.105-110.2002
Phylogeny-based rapid identification of mycoplasmas and ureaplasmas from urethritis patients
Abstract
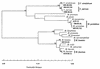
Some strains of mycoplasmas and ureaplasmas (family Mycoplasmataceae) are associated with nongonococcal urethritis (NGU) or other genitourinary infections. We have developed a rapid and reliable method of identifying the presence and prevalence of mycoplasmas and ureaplasmas in men with NGU. This method is based on the amplification of a part of the 16S rRNA gene by PCR and phylogenetic analysis. A portion of the 16S rRNA gene from 15 prototype strains was amplified with a set of common primers, and their nucleotides were sequenced. The nucleotide sequence of the V4 and V5 regions was analyzed by the neighbor-joining method. The 15 prototype strains were grouped into three distinct clusters, allowing us to clearly segregate the strains into distinct lineages. To determine the prevalence of these pathogens among patients with NGU, this protocol was tested with 148 urine samples. Amplifications were observed for 42 samples, and their nucleotide sequences were analyzed along with those of the 15 prototype strains. The phylogenetic tree thus constructed indicated that 15 of the 42 formed a cluster with Mycoplasma genitalium. Among the remaining specimens, 2 formed a cluster with Mycoplasma hominis, 19 with Ureaplasma urealyticum, and 5 with Ureaplasma parvum; the remaining sample contained both M. genitalium and U. urealyticum. This phylogeny-based identification of mycoplasmas and ureaplasmas provides not only a powerful tool for rapid diagnosis but also the basis for etiological studies of these pathogens.
Figures


References
-
- Artiushin, S., M. Duvall, and F. C. Minion. 1995. Phylogenetic analysis of mycoplasma strain ISM1499 and its assignment to the Acholeplasma oculi strain cluster. Int. J. Syst. Bacteriol. 45:104–109. - PubMed
-
- Behbahani, N., A. Blanchard, G. H. Cassell, and L. Montagnier. 1993. Phylogenetic analysis of Mycoplasma penetrans, isolated from HIV-infected patients. FEMS Microbiol. Lett. 109:63–66. - PubMed
-
- Christiansen, C., F. T. Black, and E. A. Freundt. 1981. Hybridization experiments with deoxyribonucleic acid from Ureaplasma urealyticum serovars I to III. Int. J. Syst. Bacteriol. 31:259–262.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

