Pandemic spread of cholera: genetic diversity and relationships within the seventh pandemic clone of Vibrio cholerae determined by amplified fragment length polymorphism
- PMID: 11773113
- PMCID: PMC120103
- DOI: 10.1128/JCM.40.1.172-181.2002
Pandemic spread of cholera: genetic diversity and relationships within the seventh pandemic clone of Vibrio cholerae determined by amplified fragment length polymorphism
Abstract
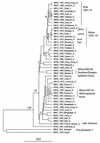
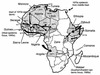
The seventh cholera pandemic started in 1961 and continues today. A collection of 45 seventh pandemic isolates of V. cholerae sampled over a 33-year period were analyzed by amplified fragment length polymorphism (AFLP) fingerprinting. All but four pairs and one set of three isolates were distinguished. AFLP revealed far more variation than ribotyping, which was until now the most useful method of revealing variation within the pandemic clone. Unfortunately, the ribotype variation observed is mainly due to recombination between the multiple copies of the rrn genes (R. Lan and P. R. Reeves, Microbiology 144:1213-1221, 1998), which makes changes susceptible to repeat occurrences and reversion. This AFLP study shows that particularly for the common ribotypes G and H, such events have indeed occurred. AFLP grouped most of the 45 isolates into two clusters. Cluster I consists mainly of strains from the 1960s and 1970s, while cluster II contains mainly strains from the 1980s and 1990s, revealing a temporal pattern of change in the clone. This is best seen in the relationships of the strains from Africa, which correlate with the epidemiology of epidemics on that continent. The data confirm independent introductions to Africa during the 1970s outbreak and reveal several other African introductions. In the 1991 cholera upsurge, isolates from the Southern and Eastern African epidemic focus are markedly different from those from the West African epidemic focus. An isolate from 1987 in Algeria was identical to the West epidemic isolates, suggesting that the strain was present in Africa at least 3 years before causing large outbreaks. These observations have major implications for our understanding of cholera epidemiology.
Figures
References
-
- Aarts, H. J., L. A. van Lith, and J. Keijer. 1998. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett. Appl. Microbiol. 26:131–135. - PubMed
-
- Barua, D. 1992. History of cholera, p.1–36. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum Press, New York, N.Y.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

