Molecular identification of enterovirus by analyzing a partial VP1 genomic region with different methods
- PMID: 11773114
- PMCID: PMC120085
- DOI: 10.1128/JCM.40.1.182-192.2002
Molecular identification of enterovirus by analyzing a partial VP1 genomic region with different methods
Abstract
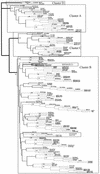
VP1 is the most suitable region for use in the identification of enterovirus. Although VP1 sequencing methods may vary, it is necessary to agree on a common strategy of sequence analysis. Identification of a strain type may be achieved by three different approaches: pairwise sequence alignment, multiple-sequence alignment, and phylogenetic inference. Other methods are also available, but they are not simple enough to be performed at a virology laboratory. The performances of these methods were evaluated with nucleotide and protein sequences obtained from 32 original samples, 8 enterovirus isolates, and 64 GenBank sequences. Pairwise sequence alignment methods had very different results. The DNASTAR package identified only 28.8% of enterovirus strains, while the Genetics Computer Group package identified 50.0 or 72.1% of enterovirus strains when nucleotide or amino acid sequences were analyzed, respectively. Multiple-sequence alignment methods identified 94.2% (Clustal W program) or 92.3% (Pileup program) of the enterovirus strains, while the phylogenetic method increased this rate to 99.0%. Comparative evaluation of these analysis methods showed that the Clustal W program (version 1.81), a freely available multiple-sequence alignment program, presented one of the best performances when used with the correct criteria. Other commercial and expensive programs did not achieve the same performances, making them less suitable for molecular typing of enteroviruses. Finally, although phylogenetic inference is the most demanding method in terms of knowledge of the user, it remained the best option analyzed.
Figures

References
-
- Barton, G. 1990. Protein multiple sequence alignment and flexible pattern matching. Methods Enzymol. 183:403–428. - PubMed
-
- Burland, T. G. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 132:71–91. - PubMed
-
- Casas, I., P. E. Klapper, G. M. Cleator, J. E. Echevarría, A. Tenorio, and J. M. Echevarría. 1995. Two different PCR assays to detect enteroviral RNA in CSF samples from patients with acute aseptic meningitis. J. Med. Virol. 47:378–385. - PubMed
-
- Casas, I. P., G. Trallero, D. Cisterna, M. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138–148. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

