Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR
- PMID: 11773402
- PMCID: PMC135859
- DOI: 10.1128/jvi.76.3.1265-1272.2002
Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR
Abstract
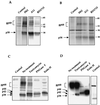
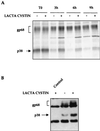
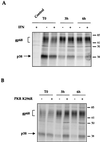
The hepatitis C virus (HCV) envelope protein E2 has been shown to accumulate in the lumen of the endoplasmic reticulum (ER) as a properly folded glycoprotein as well as large aggregates of misfolded proteins. In the present study, we have identified an additional unglycosylated species, with an apparent molecular mass of 38 kDa (E2-p38). In contrast to the glycosylated E2, E2-p38 is significantly less stable and is degraded through the proteasome pathway. Correspondingly, E2-p38 is found to be ubiquitinated. E2-p38 is localized mostly in the cytosol, in contrast to the glycosylated form, which is exclusively membrane associated. Alpha interferon (IFN-alpha) treatment or overexpression of the double-stranded RNA-activated protein kinase (PKR) significantly increased the stability of E2-p38, consistent with a previous report (D. R. Taylor, S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai, Science 285:107-110, 1999) that E2 interacts with PKR and inhibits its kinase activity. Direct interaction between PKR and E2-p38, but not the glycosylated form of E2, was also observed. These results show that E2-p38 is the form of E2 that interacts with PKR in the cytosol and may contribute to the resistance of HCV to IFN-alpha. Thus, an ER protein can exist in the cytosol as an unglycosylated species and impair cellular functions.
Figures



References
-
- Colombo, M. 1998. The role of hepatitis C virus in hepatocellular carcinoma. Recent Results Cancer Res. 154:337–344. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

