The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting
- PMID: 11773403
- PMCID: PMC135861
- DOI: 10.1128/jvi.76.3.1273-1284.2002
The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting
Abstract
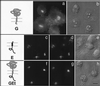
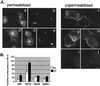
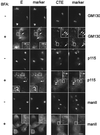
We have previously shown that the E protein of the coronavirus infectious bronchitis virus (IBV) is localized to the Golgi complex when expressed exogenously from cDNA. Here, we report that neither the transmembrane domain nor the short lumenal domain of IBV E is required for Golgi targeting. However, an N-terminal truncation containing only the cytoplasmic domain (CTE) was efficiently localized to the Golgi complex, and this domain could retain a reporter protein in the Golgi. Thus, the cytoplasmic tail of the E protein is necessary and sufficient for Golgi targeting. The IBV E protein is palmitoylated on one or two cysteine residues adjacent to its transmembrane domain, but palmitoylation was not required for proper Golgi targeting. Using C-terminal truncations, we determined that the IBV E Golgi targeting information is present between tail amino acids 13 and 63. Upon treatment with brefeldin A, both the E and CTE proteins redistributed to punctate structures that colocalized with the Golgi matrix proteins GM130 and p115 instead of being localized to the endoplasmic reticulum like Golgi glycosylation enzymes. This suggests that IBV E is associated with the Golgi matrix through interactions of its cytoplasmic tail and may have interesting implications for coronavirus assembly in early Golgi compartments.
Figures



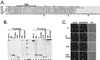


References
-
- Barr, F. A., M. Puype, J. Vandekerckhove, and G. Warren. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91:253–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

