The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion
- PMID: 11773407
- PMCID: PMC135857
- DOI: 10.1128/jvi.76.3.1328-1338.2002
The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion
Abstract
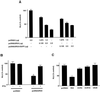
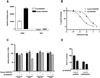
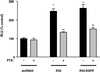
The rat cytomegalovirus (RCMV) R33 gene is conserved among all betaherpesviruses and encodes a protein (pR33) that shows sequence similarity with chemokine-binding G protein-coupled receptors (GPCRs). Previously, the physiological significance of the R33 gene was demonstrated by the finding that an RCMV strain with R33 deleted is severely attenuated in vivo and is unable to either enter or replicate in the salivary glands of infected rats. Here, we report that RCMV pR33 is expressed as a functional GPCR that signals in an agonist-independent manner in both COS-7 and Rat2 cells. Transient expression of pR33 in COS-7 cells results in constitutive activation of phospholipase C (PLC) due to coupling to G proteins of the G(q) class. Interestingly, PLC activation is partially inhibited by cotransfection with G(alpha)-transducin subunits, which indicates the involvement of G(betagamma) as well as Galpha subunits in pR33-mediated signaling. Surprisingly, PLC activation is also partially inhibited by addition of pertussis toxin (PTX), suggesting that pR33 activates not only G(q) but also G(i/0) proteins. The constitutive activation of G(i/0) proteins by pR33 is further demonstrated by the PTX-sensitive decrease of CRE-mediated transcription and the PTX-sensitive increase of both NF-kappaB- and SRE-mediated transcription. In contrast to its homolog of human herpesvirus 6B (pU12), pR33 does not bind RANTES.
Figures
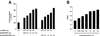
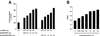
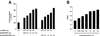
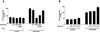
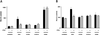
Similar articles
-
Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and Gs classes.J Biol Chem. 2003 Dec 12;278(50):50010-23. doi: 10.1074/jbc.M306530200. Epub 2003 Sep 30. J Biol Chem. 2003. PMID: 14522997
-
The rat cytomegalovirus R78 G protein-coupled receptor gene is required for production of infectious virus in the spleen.J Gen Virol. 2003 Sep;84(Pt 9):2517-2530. doi: 10.1099/vir.0.19227-0. J Gen Virol. 2003. PMID: 12917474
-
Mutational analysis of the R33-encoded G protein-coupled receptor of rat cytomegalovirus: identification of amino acid residues critical for cellular localization and ligand-independent signalling.J Gen Virol. 2004 Apr;85(Pt 4):897-909. doi: 10.1099/vir.0.19709-0. J Gen Virol. 2004. PMID: 15039532
-
Multiple pathways of ERK activation by G protein-coupled receptors.Novartis Found Symp. 2001;239:68-79; discussion 80-4, 150-9. doi: 10.1002/0470846674.ch7. Novartis Found Symp. 2001. PMID: 11529317 Review.
-
G protein specificity: traffic direction required.Cell Signal. 2002 May;14(5):407-18. doi: 10.1016/s0898-6568(01)00259-5. Cell Signal. 2002. PMID: 11882385 Review.
Cited by
-
Receptor chimeras demonstrate that the C-terminal domain of the human cytomegalovirus US27 gene product is necessary and sufficient for intracellular receptor localization.Virol J. 2012 Feb 16;9:42. doi: 10.1186/1743-422X-9-42. Virol J. 2012. PMID: 22339884 Free PMC article.
-
Cellular distribution of CD200 receptor in rats and its interaction with cytomegalovirus e127 protein.Med Microbiol Immunol. 2018 Nov;207(5-6):307-318. doi: 10.1007/s00430-018-0552-3. Epub 2018 Jul 21. Med Microbiol Immunol. 2018. PMID: 30032349
-
Role of G protein-coupled receptors in inflammation.Acta Pharmacol Sin. 2012 Mar;33(3):342-50. doi: 10.1038/aps.2011.200. Epub 2012 Feb 27. Acta Pharmacol Sin. 2012. PMID: 22367283 Free PMC article. Review.
-
The Epstein-Barr virus BILF1 gene encodes a G protein-coupled receptor that inhibits phosphorylation of RNA-dependent protein kinase.J Virol. 2005 Jan;79(1):441-9. doi: 10.1128/JVI.79.1.441-449.2005. J Virol. 2005. PMID: 15596837 Free PMC article.
-
Why Are Cytomegalovirus-Encoded G-Protein-Coupled Receptors Essential for Infection but Only Variably Conserved?Pathogens. 2025 Mar 3;14(3):245. doi: 10.3390/pathogens14030245. Pathogens. 2025. PMID: 40137730 Free PMC article. Review.
References
-
- Arai, H., and I. F. Charo. 1996. Differential regulation of G-protein-mediated signaling by chemokine receptors. J. Biol. Chem. 271:21814–21819. - PubMed
-
- Arvanitakis, L., E. Geras-Raaka, A. Varma, M. C. Gershenghorn, and E. Cesarman. 1997. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385:347–350. - PubMed
-
- Bais, C., B. Santamasso, O. Coso, L. Arvanitakis, E. Geras-Raaka, J. Silvio Gutkind, A. S. Asch, E. Cesarman, M. C. Gershenghorn, and E. A. Mesri. 1998. G-protein-coupled receptor of Kaposi’s sarcoma-associated is a viral oncogene and angiogenesis activator. Nature 391:86–89. (Erratum, 392:210.) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

