Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease
- PMID: 11773410
- PMCID: PMC135855
- DOI: 10.1128/jvi.76.3.1359-1368.2002
Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease
Abstract
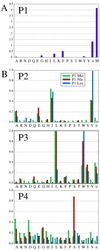
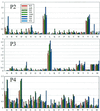
Resistance to human immunodeficiency virus type 1 protease (HIV PR) inhibitors results primarily from the selection of multiple mutations in the protease region. Because many of these mutations are selected for the ability to decrease inhibitor binding in the active site, they also affect substrate binding and potentially substrate specificity. This work investigates the substrate specificity of a panel of clinically derived protease inhibitor-resistant HIV PR variants. To compare protease specificity, we have used positional-scanning, synthetic combinatorial peptide libraries as well as a select number of individual substrates. The subsite preferences of wild-type HIV PR determined by using the substrate libraries are consistent with prior reports, validating the use of these libraries to compare specificity among a panel of HIV PR variants. Five out of seven protease variants demonstrated subtle differences in specificity that may have significant impacts on their abilities to function in viral maturation. Of these, four variants demonstrated up to fourfold changes in the preference for valine relative to alanine at position P2 when tested on individual peptide substrates. This change correlated with a common mutation in the viral NC/p1 cleavage site. These mutations may represent a mechanism by which severely compromised, drug-resistant viral strains can increase fitness levels. Understanding the altered substrate specificity of drug-resistant HIV PR should be valuable in the design of future generations of protease inhibitors as well as in elucidating the molecular basis of regulation of proteolysis in HIV.
Figures


References
-
- Backes, B. J., J. L. Harris, F. Leonetti, C. S. Craik, and J. A. Ellman. 2000. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 18:187–193. - PubMed
-
- Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retrovir. 16:1209–1213. - PubMed
-
- Beck, Z. Q., L. Hervio, P. E. Dawson, J. H. Elder, and E. L. Madison. 2000. Identification of efficiently cleaved substrates for HIV-1 protease using a phage display library and use in inhibitor development. Virology 274:391–401. - PubMed
-
- Billich, A., and G. Winkler. 1991. Analysis of subsite preferences of HIV-1 proteinase using MA/CA junction peptides substituted at the P3–P1′ positions. Arch. Biochem. Biophys. 290:186–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

