Human paralogs of KIAA0187 were created through independent pericentromeric-directed and chromosome-specific duplication mechanisms
- PMID: 11779832
- PMCID: PMC155266
- DOI: 10.1101/gr.213702
Human paralogs of KIAA0187 were created through independent pericentromeric-directed and chromosome-specific duplication mechanisms
Abstract
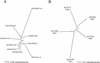
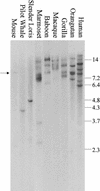
KIAA0187 is a gene of unknown function that maps to 10q11 and has been subject to recent duplication events. Here we analyze 18 human paralogs of this gene and show that paralogs of exons 14-23 were formed through satellite-associated pericentromeric-directed duplication, whereas paralogs of exons 1-9 were created via chromosome-specific satellite-independent duplications. In silico, Northern, and RT-PCR analyses indicate that nine paralogs are transcribed, including four in which KIAA0187 exons are spliced onto novel sequences. Despite this, no new genes appear to have been created by these events. The chromosome 10 paralogs map to 10q11, 10q22, 10q23.1, and 10q23.3, forming part of a complex family of chromosome-specific repeats that includes GLUD1, Cathepsin L, and KIAA1099 pseudogenes. Phylogenetic analyses and comparative FISH indicates that the 10q23.1 and 10q23.3 repeats were created in 10q11 and relocated by a paracentric inversion 13 to 27 Myr ago. Furthermore, the most recent duplications, involving the KIAA1099 pseudogenes, have largely been confined to 10q11. These results indicate a simple model for the evolution of this repeat family, involving multiple rounds of centromere-proximal duplication and dispersal through intrachromosomal rearrangement. However, more complex events must be invoked to account for high sequence identity between some paralogs.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Antonacci R, Marzella R, Finelli P, Lonoce A, Forabosco A, Archiadiacono N, Rocchi M. A panel of subchromosomal painting libraries representing over 300 regions of the human genome. Cytogenet Cell Genet. 1995;68:25–32. - PubMed
-
- Bayne RA, Broccoli D, Taggart MH, Thomson EJ, Farr CJ, Cooke HJ. Sandwiching of a gene within 12 kb of a functional telomere and alpha satellite does not result in silencing. Hum Mol Genet. 1994;3:539–546. - PubMed
-
- Barber JC, Cross IE, Douglas F, Nicholson JC, Moore KJ, Browne CE. Neurofibromatosis pseudogene amplification underlies euchromatic cytogenetic duplications and triplications of proximal 15q. Hum Genet. 1998;103:600–607. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
