Identification of the modifier of Min 2 (Mom2) locus, a new mutation that influences Apc-induced intestinal neoplasia
- PMID: 11779834
- PMCID: PMC155250
- DOI: 10.1101/gr.206002
Identification of the modifier of Min 2 (Mom2) locus, a new mutation that influences Apc-induced intestinal neoplasia
Abstract
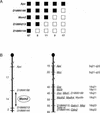
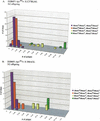
Min (Multiple intestinal neoplasia) mice carry a dominant mutation in the adenomatous polyposis coli (Apc) gene and develop multiple adenomas throughout their intestinal tract (Moser et al. 1990; Su et al 1992). Polyp multiplicity in Min mice is greatly influenced by genetic background. A modifier locus, Mom1 (Modifier of Min 1), was identified and localized to distal mouse chromosome 4 (Moser et al. 1992; Dietrich et al. 1993), and accounts for some of the genetic variance in polyp multiplicity. Mom1 is a semidominant modifier of polyp size and multiplicity in Min mice (Gould and Dove 1997), and encodes the secretory type II nonpancreatic phospholipase A2 (Pla2g2a) gene (MacPhee et al. 1995; Cornier et al. 1997, 2000). We now report the identification of a second Modifier of Min 2 (Mom2) locus that is the result of a spontaneous mutation. One resistant Mom2 allele can suppress 88%-95% of polyps detected in Apc(Min)/+ mice, indicating that Mom2 acts in a dominant fashion. Linkage analysis has localized Mom2 to distal mouse chromosome 18. The effects of the Mom2 locus on reducing polyp multiplicity are stronger than the effects of the Mom1 locus, in both the small and large intestines. Some Apc(Min)/+ mice that carried one resistant Mom2 allele were tumor-free at 21 weeks of age, even in the absence of a resistant Mom1 allele. Thus, the resistant Mom2 allele can, in some cases, completely suppress the penetrance of the Apc(Min) mutation.
Figures

Similar articles
-
The modifier of Min 2 (Mom2) locus: embryonic lethality of a mutation in the Atp5a1 gene suggests a novel mechanism of polyp suppression.Genome Res. 2007 May;17(5):566-76. doi: 10.1101/gr.6089707. Epub 2007 Mar 26. Genome Res. 2007. PMID: 17387143 Free PMC article.
-
The CAST/Ei strain confers significant protection against Apc(Min) intestinal polyps, independent of the resistant modifier of Min 1 (Mom1) locus.Cancer Res. 2002 Oct 1;62(19):5413-7. Cancer Res. 2002. PMID: 12359746
-
Analysis of reciprocal congenic lines reveals the C3H/HeJ genome to be highly resistant to ApcMin intestinal tumorigenesis.Genomics. 2004 Nov;84(5):844-52. doi: 10.1016/j.ygeno.2004.07.010. Genomics. 2004. PMID: 15475263
-
The intestinal epithelium and its neoplasms: genetic, cellular and tissue interactions.Philos Trans R Soc Lond B Biol Sci. 1998 Jun 29;353(1370):915-23. doi: 10.1098/rstb.1998.0256. Philos Trans R Soc Lond B Biol Sci. 1998. PMID: 9684289 Free PMC article. Review.
-
Analysis of the Mom1 modifier of intestinal neoplasia in mice.Exp Lung Res. 1998 Jul-Aug;24(4):437-53. doi: 10.3109/01902149809087379. Exp Lung Res. 1998. PMID: 9659576 Review.
Cited by
-
The modifier of Min 2 (Mom2) locus: embryonic lethality of a mutation in the Atp5a1 gene suggests a novel mechanism of polyp suppression.Genome Res. 2007 May;17(5):566-76. doi: 10.1101/gr.6089707. Epub 2007 Mar 26. Genome Res. 2007. PMID: 17387143 Free PMC article.
-
Identification of Mom12 and Mom13, two novel modifier loci of Apc (Min) -mediated intestinal tumorigenesis.Cell Cycle. 2011 Apr 1;10(7):1092-9. doi: 10.4161/cc.10.7.15089. Epub 2011 Apr 1. Cell Cycle. 2011. PMID: 21386660 Free PMC article.
-
Understanding phenotypic variation in rodent models with germline Apc mutations.Cancer Res. 2013 Apr 15;73(8):2389-99. doi: 10.1158/0008-5472.CAN-12-4607. Epub 2013 Apr 11. Cancer Res. 2013. PMID: 23580574 Free PMC article. Review.
-
Cables1 is a tumor suppressor gene that regulates intestinal tumor progression in Apc(Min) mice.Cancer Biol Ther. 2013 Jul;14(7):672-8. doi: 10.4161/cbt.25089. Epub 2013 May 31. Cancer Biol Ther. 2013. PMID: 23792637 Free PMC article.
-
Novel murine mammary epithelial cell lines that form osteolytic bone metastases: effect of strain background on tumor homing.Clin Exp Metastasis. 2003;20(2):111-20. doi: 10.1023/a:1022675031185. Clin Exp Metastasis. 2003. PMID: 12705632
References
-
- Balmain A, Nagase H. Cancer resistance genes in mice: Models for the study of tumour modifiers. Trends Genet. 1998;14:139–144. - PubMed
-
- Bedell MA, Largaespada DA, Jenkins NA, Copeland NG. Mouse models of human disease. Part II: Recent progress and future directions. Genes Dev. 1997;11:11–43. - PubMed
-
- Bocker T, Ruschoff J, Fishel R. Molecular diagnostics of cancer predisposition: Hereditary non-polyposis colorectal carcinoma and mismatch repair defects. Biochim Biophys Acta. 1999;1423:O1–O10. - PubMed
-
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. - PubMed
-
- Buchberg AM, Siracusa LD. The Genetics of Cancer Susceptibility. In: Fortner JG, Rhoads JE, editors. General Motors Cancer Research Foundation: Accomplishments in Cancer Research. Philadelphia, PA: J.B. Lippincott Company; 1995.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
