Changes in gene expression profiles in developing B cells of murine bone marrow
- PMID: 11779835
- PMCID: PMC155249
- DOI: 10.1101/gr.201501
Changes in gene expression profiles in developing B cells of murine bone marrow
Abstract
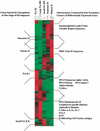
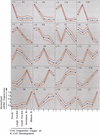
Gene expression profiles of five consecutive stages of mouse B cell development were generated with high-density oligonucleotide arrays from as few as 2 x 10(4) ex vivo isolated and flow-cytometrically purified cells. Between 2.8% and 6.8% of all genes change on differentiation from one cellular stage to the next by at least twofold. The entire pathway involves differential expression of 10.7% of all genes. Previously known expression patterns of 15 genes (like surrogate light chain, RAG-1/2, MHC class II, mel-14 antigen) are confirmed. The gene expression patterns of the proliferating pre-BI and large pre-BII cells on the one hand, and the resting immature and mature B cells on the other hand, are most similar to each other. Small pre-BII cells display a pattern that is transitional between these two groups. Most of the genes expressed in early precursors are involved in general processes, like protein folding or cell cycle regulation, whereas more mature precursors express genes involved in more specific molecular programs (cell surface receptors, secreted factors, and adhesion molecules, among others). Between 19 and 139 genes share a given expression pattern. Combining knowledge about gene function and expression pattern allows identification of novel candidate genes potentially involved in self-maintenance of pre-BI cells, allelic exclusion and pre-B cell receptor signaling in large pre BII cells, cell-cycle arrest of small pre-BII cells, propensity toward apoptosis or anergization in immature B cells, propensity toward cell division and activation in mature B cells, and stage-specific interactions with stromal cells in the bone marrow.
Figures


References
-
- Cheng G, Baltimore D. TANK, a co-inducer with TRAF2 of TNF- and CD 40L-mediated NF-kappaB activation. Genes & Dev. 1996;10:963–973. - PubMed
-
- Chin D, Means AR. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. - PubMed
-
- Coutavas EE, Hsieh CM, Ren M, Drivas GT, Rush MG, D'Eustachio PD. Tissue-specific expression of Ran isoforms in the mouse. Mamm Genome. 1994;5:623–628. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
