The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus
- PMID: 11781357
- PMCID: PMC150824
- DOI: 10.1172/JCI14080
The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus
Abstract
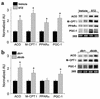
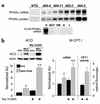
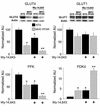
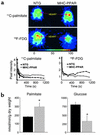
Recent evidence has defined an important role for PPARalpha in the transcriptional control of cardiac energy metabolism. To investigate the role of PPARalpha in the genesis of the metabolic and functional derangements of diabetic cardiomyopathy, mice with cardiac-restricted overexpression of PPARalpha (MHC-PPAR) were produced and characterized. The expression of PPARalpha target genes involved in cardiac fatty acid uptake and oxidation pathways was increased in MHC-PPAR mice. Surprisingly, the expression of genes involved in glucose transport and utilization was reciprocally repressed in MHC-PPAR hearts. Consistent with the gene expression profile, myocardial fatty acid oxidation rates were increased and glucose uptake and oxidation decreased in MHC-PPAR mice, a metabolic phenotype strikingly similar to that of the diabetic heart. MHC-PPAR hearts exhibited signatures of diabetic cardiomyopathy including ventricular hypertrophy, activation of gene markers of pathologic hypertrophic growth, and transgene expression-dependent alteration in systolic ventricular dysfunction. These results demonstrate that (a) PPARalpha is a critical regulator of myocardial fatty acid uptake and utilization, (b) activation of cardiac PPARalpha regulatory pathways results in a reciprocal repression of glucose uptake and utilization pathways, and (c) derangements in myocardial energy metabolism typical of the diabetic heart can become maladaptive, leading to cardiomyopathy.
Figures

References
-
- Kannel WB, Hjortland M, Castelli WP. Role of diabetes in congestive heart failure: the Framingham study. Am J Cardiol. 1974; 34:29–34. - PubMed
-
- Rubler S, et al. New type of cardiomyopathy associated with glomerulosclerosis. Am J Cardiol. 1972; 30:595–602. - PubMed
-
- Stanley WC, Lopaschuk GD, McCormack JG. Regulation of energy substrate metabolism in the diabetic heart. Cardiovasc Res. 1997; 34:25–33. - PubMed
-
- Rodrigues B, Cam MC, McNeill JH. Myocardial substrate metabolism: implications for diabetic cardiomyopathy. J Mol Cell Cardiol. 1995; 27:169–179. - PubMed
-
- Neely JR, Rovetto MJ, Oram JF. Myocardial utilization of carbohydrate and lipids. Prog Cardiovasc Dis. 1972; 15:289–329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 HL13851/HL/NHLBI NIH HHS/United States
- JDFI 996003/PHS HHS/United States
- P01 HL5727805/HL/NHLBI NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- P30 DK56341/DK/NIDDK NIH HHS/United States
- R01 DK045416/DK/NIDDK NIH HHS/United States
- P01 HL013851/HL/NHLBI NIH HHS/United States
- F32 HL067575/HL/NHLBI NIH HHS/United States
- P30 DK52574/DK/NIDDK NIH HHS/United States
- F32 HL67575/HL/NHLBI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- R01 DK45416/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

