Plasmodium falciparum phospholipase C hydrolyzing sphingomyelin and lysocholinephospholipids is a possible target for malaria chemotherapy
- PMID: 11781362
- PMCID: PMC2196011
- DOI: 10.1084/jem.20010724
Plasmodium falciparum phospholipase C hydrolyzing sphingomyelin and lysocholinephospholipids is a possible target for malaria chemotherapy
Abstract
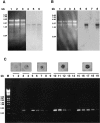
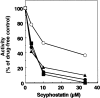
Sphingomyelinase (SMase) is one of the principal enzymes in sphingomyelin (SM) metabolism. Here, we identified a Plasmodium falciparum gene (PfNSM) encoding a 46-kD protein, the amino acid sequence of which is approximately 25% identical to that of bacteria SMases. Biochemical analyses of the recombinant protein GST-PfNSM, a fusion protein of the PfNSM product with glutathione-S-transferase, reveal that this enzyme retained similar characteristics in various aspects to SMase detected in P. falciparum-infected erythrocytes and isolated parasites. In addition, the recombinant protein retains hydrolyzing activity not only of SM but also of lysocholinephospholipids (LCPL) including lysophosphatidylcholine and lysoplatelet-activating factor, indicating that PfNSM encodes SM/LCPL-phospholipase C (PLC). Scyphostatin inhibited SM/LCPL-PLC activities of the PfNSM product as well as the intraerythrocytic proliferation of P. falciparum in a dose-dependent manner with ID(50) values for SM/LCPL-PLC activities and the parasite growth at 3-5 microM and approximately 7 microM, respectively. Morphological analysis demonstrated most severe impairment in the intraerythrocytic development with the addition of scyphostatin at trophozoite stage than at ring or schizont stages, suggesting its effect specifically on the stage progression from trophozoite to schizont, coinciding with the active transcription of PfNSM gene.
Figures


Similar articles
-
Neutral sphingomyelinase activity dependent on Mg2+ and anionic phospholipids in the intraerythrocytic malaria parasite Plasmodium falciparum.Biochem J. 2000 Mar 15;346 Pt 3(Pt 3):671-7. Biochem J. 2000. PMID: 10698693 Free PMC article.
-
Computational identification and experimental characterization of substrate binding determinants of nucleotide pyrophosphatase/phosphodiesterase 7.BMC Biochem. 2011 Dec 16;12:65. doi: 10.1186/1471-2091-12-65. BMC Biochem. 2011. PMID: 22177013 Free PMC article.
-
Function of the cloned putative neutral sphingomyelinase as lyso-platelet activating factor-phospholipase C.J Biol Chem. 1999 Dec 31;274(53):38131-9. doi: 10.1074/jbc.274.53.38131. J Biol Chem. 1999. PMID: 10608884
-
Plasmodium falciparum glycogen synthase kinase-3: molecular model, expression, intracellular localisation and selective inhibitors.Biochim Biophys Acta. 2004 Mar 11;1697(1-2):181-96. doi: 10.1016/j.bbapap.2003.11.023. Biochim Biophys Acta. 2004. PMID: 15023360 Review.
-
Membrane fusion induced by phospholipase C and sphingomyelinases.Biosci Rep. 2000 Dec;20(6):443-63. doi: 10.1023/a:1010450702670. Biosci Rep. 2000. PMID: 11426688 Review.
Cited by
-
PG12, a phospholipid analog with potent antimalarial activity, inhibits Plasmodium falciparum CTP:phosphocholine cytidylyltransferase activity.J Biol Chem. 2011 Aug 19;286(33):28940-28947. doi: 10.1074/jbc.M111.268946. Epub 2011 Jun 25. J Biol Chem. 2011. PMID: 21705805 Free PMC article.
-
Identification of Sphingomyelinase on the Surface of Chlamydia pneumoniae: Possible Role in the Entry into Its Host Cells.Interdiscip Perspect Infect Dis. 2014;2014:412827. doi: 10.1155/2014/412827. Epub 2014 Mar 13. Interdiscip Perspect Infect Dis. 2014. PMID: 24757444 Free PMC article.
-
Acyclic immucillin phosphonates: second-generation inhibitors of Plasmodium falciparum hypoxanthine-guanine-xanthine phosphoribosyltransferase.Chem Biol. 2012 Jun 22;19(6):721-30. doi: 10.1016/j.chembiol.2012.04.012. Chem Biol. 2012. PMID: 22726686 Free PMC article.
-
Enzymatic Transition States and Drug Design.Chem Rev. 2018 Nov 28;118(22):11194-11258. doi: 10.1021/acs.chemrev.8b00369. Epub 2018 Oct 18. Chem Rev. 2018. PMID: 30335982 Free PMC article. Review.
-
Anti-Plasmodium activity of ceramide analogs.Malar J. 2004 Dec 10;3:49. doi: 10.1186/1475-2875-3-49. Malar J. 2004. PMID: 15588325 Free PMC article.
References
-
- van Deenen, L.L.M., and J. de Gier. 1974. Lipids of the red cell membrane. The Red Blood Cell. D.M. Surgenor, editor. Academic Press, New York. 147–211.
-
- Atkinson, C.T., and M. Aikawa. 1990. Ultrastructure of malaria-infected erythrocytes. Blood Cells. 16:351–368. - PubMed
-
- Vial, H.J., and M.L. Ancelin. 1998. Malarial lipids. Malaria: Parasite Biology, Pathogenesis, and Protection. I.W. Sherman, editor. ASM Press, Washington, D.C. 159–175.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

