CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation
- PMID: 11782425
- PMCID: PMC125809
- DOI: 10.1093/emboj/21.1.53
CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation
Abstract
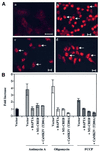
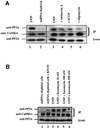
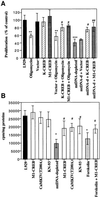
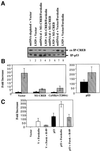
We characterized a new signaling pathway leading to the activation of cAMP-responsive element-binding protein (CREB) in several cell lines affected by mitochondrial dysfunction. In vitro kinase assays, inhibitors of several kinase pathways and overexpression of a dominant-negative mutant for calcium/calmodulin kinase IV (CaMKIV), which blocks the activation of CREB, showed that CaMKIV is activated by a mitochondrial activity impairment. A high calcium concentration leading to the disruption of the protein interaction with protein phosphatase 2A explains CaMKIV activation in these conditions. Transcrip tionally active phosphorylated CREB was also found in a rho0 143B human osteosarcoma cell line and in a MERRF cybrid cell line mutated for tRNA(Lys) (A8344G). We also showed that phosphorylated CREB is involved in the proliferation defect induced by a mitochondrial dysfunction. Indeed, cell proliferation inhibition can be prevented by CaMKIV inhibition and CREB dominant-negative mutants. Finally, our data suggest that phosphorylated CREB recruits p53 tumor suppressor protein, modifies its transcriptional activity and increases the expression of p21(Waf1/Cip1), a p53-regulated cyclin-dependent kinase inhibitor.
Figures

Similar articles
-
Regulation and function of the calcium/calmodulin-dependent protein kinase IV/protein serine/threonine phosphatase 2A signaling complex.J Biol Chem. 2004 Jul 23;279(30):31708-16. doi: 10.1074/jbc.M404523200. Epub 2004 May 13. J Biol Chem. 2004. PMID: 15143065
-
A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A.Science. 1998 May 22;280(5367):1258-61. doi: 10.1126/science.280.5367.1258. Science. 1998. PMID: 9596578
-
Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity.Genes Dev. 1994 Nov 1;8(21):2527-39. doi: 10.1101/gad.8.21.2527. Genes Dev. 1994. PMID: 7958915
-
Regulation and properties of the rat Ca2+/calmodulin-dependent protein kinase IV gene and its protein products.Recent Prog Horm Res. 1997;52:389-406; discussion 406-7. Recent Prog Horm Res. 1997. PMID: 9238860 Review.
-
Ca2+/calmodulin-dependent protein kinase IV and calcium signaling.Biometals. 1998 Dec;11(4):331-43. doi: 10.1023/a:1009276932076. Biometals. 1998. PMID: 10191497 Review.
Cited by
-
A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation.Protein Cell. 2015 Dec;6(12):862-70. doi: 10.1007/s13238-015-0175-z. Epub 2015 Jun 18. Protein Cell. 2015. PMID: 26084519 Free PMC article. Review.
-
CREB: A Key Regulator of Normal and Neoplastic Hematopoiesis.Adv Hematol. 2009;2009:634292. doi: 10.1155/2009/634292. Epub 2009 Aug 27. Adv Hematol. 2009. PMID: 19960054 Free PMC article.
-
Mitochondrial localization, import, and mitochondrial function of the androgen receptor.J Biol Chem. 2019 Apr 19;294(16):6621-6634. doi: 10.1074/jbc.RA118.006727. Epub 2019 Feb 21. J Biol Chem. 2019. PMID: 30792308 Free PMC article.
-
HnRNPA2 is a novel histone acetyltransferase that mediates mitochondrial stress-induced nuclear gene expression.Cell Discov. 2016 Dec 6;2:16045. doi: 10.1038/celldisc.2016.45. eCollection 2016. Cell Discov. 2016. PMID: 27990297 Free PMC article.
-
DNA damage related crosstalk between the nucleus and mitochondria.Free Radic Biol Med. 2017 Jun;107:216-227. doi: 10.1016/j.freeradbiomed.2016.11.050. Epub 2016 Nov 30. Free Radic Biol Med. 2017. PMID: 27915046 Free PMC article. Review.
References
-
- Biswas G., Adebanjo,O.A., Freedman,B.D., Anandatheerthavarada,H.K., Vijayasarathy,C., Zaidi,M., Kotlikoff,M. and Avadhani,N.G. (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J., 18, 522–533. - PMC - PubMed
-
- Bonni A., Brunet,A., West,A.E., Datta,S.R., Takasu,M.A. and Greenberg,M.E. (1999) Cell survival promoted by the Ras–MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science, 286, 1358–1362. - PubMed
-
- Buchet K. and Godinot,C. (1998) Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted ρ° cells. J. Biol. Chem., 273, 22983–22989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

