Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides
- PMID: 11782528
- PMCID: PMC117354
- DOI: 10.1073/pnas.012583999
Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides
Abstract
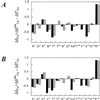
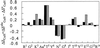
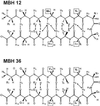
Here we present a combinatorial approach to evolve a stable beta-hairpin fold in a linear peptide. Starting with a de novo-designed linear peptide that shows a beta-hairpin structure population of around 30%, we selected four positions to build up a combinatorial library of 20(4) sequences. Deconvolution of the library using circular dichroism reduced such a sequence complexity to 36 defined sequences. Circular dichroism and NMR of these peptides resulted in the identification of two linear 14-aa-long peptides that in plain buffered solutions showed a percentage of beta-hairpin structure higher than 70%. Our results show how combinatorial approaches can be used to obtain highly structured peptide sequences that could be used as templates in which functionality can be introduced.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

