Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast
- PMID: 11782531
- PMCID: PMC117360
- DOI: 10.1073/pnas.022373099
Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast
Abstract
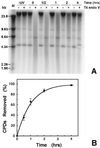
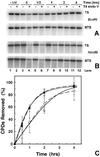
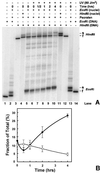
Nucleotide excision repair (NER) of UV-induced cyclobutane pyrimidine dimers (CPDs) was measured in the individual strands of transcriptionally active and inactive ribosomal genes of yeast. Ribosomal genes (rDNA) are present in multiple copies, but only a fraction of them is actively transcribed. Restriction enzyme digestion was used to specifically release the transcriptionally active fraction from yeast nuclei, and selective psoralen crosslinking was used to distinguish between active and inactive rDNA chromatin. Removal of CPDs was followed in both rDNA populations, and the data clearly show that strand-specific repair occurs in transcriptionally active rDNA while being absent in the inactive rDNA fraction. Thus, transcription-coupled repair occurs in RNA polymerase I-transcribed genes in yeast. Moreover, the nontranscribed strand of active rDNA is repaired faster than either strand of inactive rDNA, implying that NER has preferred access to the active, non-nucleosomal rDNA chromatin. Finally, restriction enzyme accessibility to active rDNA varies during NER, suggesting that there is a change in ribosomal gene chromatin structure during or soon after CPD removal.
Figures

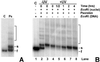

References
-
- Freidberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol. Press; 1995.
-
- Woodgate R. Genes Dev. 1999;13:2191–2195. - PubMed
-
- Vrieling H, van Zeeland A A, Mullenders L H. Mutat Res. 1998;400:135–142. - PubMed
-
- Tornaletti S, Hanawalt P C. Biochimie. 1999;81:139–146. - PubMed
-
- Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

