Improved recognition of native-like protein structures using a family of designed sequences
- PMID: 11782533
- PMCID: PMC117367
- DOI: 10.1073/pnas.022408799
Improved recognition of native-like protein structures using a family of designed sequences
Abstract
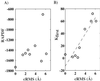
The goal of the inverse protein folding problem is to identify amino acid sequences that stabilize a given target protein conformation. Methods that attempt to solve this problem have proven useful for protein sequence design. Here we show that the same methods can provide valuable information for protein fold recognition and for ab initio protein structure prediction. We present a measure of the compatibility of a test sequence with a target model structure, based on computational protein design. The model structure is used as input to design a family of low free energy sequences, and these sequences are compared with the test sequence by using a metric in sequence space based on nearest-neighbor connectivity. We find that this measure is able to recognize the native fold of a myoglobin sequence among different globin folds. It is also powerful enough to recognize near-native protein structures among non-native models.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

