Translocation after synthesis of a four-nucleotide RNA commits RNA polymerase II to promoter escape
- PMID: 11784853
- PMCID: PMC133543
- DOI: 10.1128/MCB.22.3.762-773.2002
Translocation after synthesis of a four-nucleotide RNA commits RNA polymerase II to promoter escape
Abstract
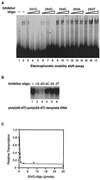
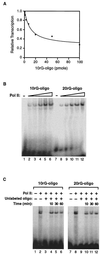
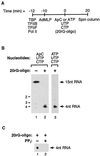
Transcription is a complex process, the regulation of which is crucial for cellular and organismic growth and development. Deciphering the molecular mechanisms that define transcription is essential to understanding the regulation of RNA synthesis. Here we describe the molecular mechanism of escape commitment, a critical step in early RNA polymerase II transcription. During escape commitment ternary transcribing complexes become stable and committed to proceeding forward through promoter escape and the remainder of the transcription reaction. We found that the point in the transcription reaction at which escape commitment occurs depends on the length of the transcript RNA (4 nucleotides [nt]) as opposed to the position of the active site of the polymerase with respect to promoter DNA elements. We found that single-stranded nucleic acids can inhibit escape commitment, and we identified oligonucleotides that are potent inhibitors of this specific step. These inhibitors bind RNA polymerase II with low nanomolar affinity and sequence specificity, and they block both promoter-dependent and promoter-independent transcription, the latter occurring in the absence of general transcription factors. We demonstrate that escape commitment involves translocation of the RNA polymerase II active site between synthesis of the third and fourth phosphodiester bonds. We propose that a conformational change in ternary transcription complexes occurs during translocation after synthesis of a 4-nt RNA to render complexes escape committed.
Figures






Similar articles
-
Kinetic and mechanistic analysis of the RNA polymerase II transcrption reaction at the human interleukin-2 promoter.J Mol Biol. 2001 Dec 14;314(5):993-1006. doi: 10.1006/jmbi.2000.5215. J Mol Biol. 2001. PMID: 11743717
-
Promoter escape by RNA polymerase II. Downstream promoter DNA is required during multiple steps of early transcription.J Biol Chem. 2003 Mar 21;278(12):10250-6. doi: 10.1074/jbc.M210848200. Epub 2003 Jan 13. J Biol Chem. 2003. PMID: 12527757
-
Promoter escape by RNA polymerase II. A role for an ATP cofactor in suppression of arrest by polymerase at promoter-proximal sites.J Biol Chem. 1996 Sep 20;271(38):23352-6. doi: 10.1074/jbc.271.38.23352. J Biol Chem. 1996. PMID: 8798537
-
Promoter escape by RNA polymerase II.Biochim Biophys Acta. 2002 Sep 13;1577(2):208-223. doi: 10.1016/s0167-4781(02)00453-0. Biochim Biophys Acta. 2002. PMID: 12213653 Review.
-
Promoter clearance and escape in prokaryotes.Biochim Biophys Acta. 2002 Sep 13;1577(2):191-207. doi: 10.1016/s0167-4781(02)00452-9. Biochim Biophys Acta. 2002. PMID: 12213652 Review.
Cited by
-
An 8 nt RNA triggers a rate-limiting shift of RNA polymerase II complexes into elongation.EMBO J. 2006 Jul 12;25(13):3100-9. doi: 10.1038/sj.emboj.7601197. Epub 2006 Jun 15. EMBO J. 2006. PMID: 16778763 Free PMC article.
-
RNA polymerase II complexes in the very early phase of transcription are not susceptible to TFIIS-induced exonucleolytic cleavage.Nucleic Acids Res. 2002 Jun 1;30(11):2290-8. doi: 10.1093/nar/30.11.2290. Nucleic Acids Res. 2002. PMID: 12034815 Free PMC article.
-
Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription.RNA. 2007 Apr;13(4):583-96. doi: 10.1261/rna.310307. Epub 2007 Feb 16. RNA. 2007. PMID: 17307818 Free PMC article.
-
The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23.Proc Natl Acad Sci U S A. 2003 May 13;100(10):5700-5. doi: 10.1073/pnas.1037057100. Epub 2003 Apr 28. Proc Natl Acad Sci U S A. 2003. PMID: 12719526 Free PMC article.
-
Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation.Biochim Biophys Acta. 2011 Jan;1809(1):34-45. doi: 10.1016/j.bbagrm.2010.11.001. Epub 2010 Nov 13. Biochim Biophys Acta. 2011. PMID: 21081187 Free PMC article. Review.
References
-
- Bowser, C., and M. Hanna. 1991. Sigma subunit of Escherichia coli RNA polymerase loses contacts with the 3′ end of the nascent RNA after synthesis of a tetranucleotide. J. Mol. Biol. 220:227–239. - PubMed
-
- Cai, H., and D. S. Luse. 1987. Transcription initiation by RNA polymerase II in vitro: properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262:298–304. - PubMed
-
- Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. - PubMed
-
- Cheetham, G. M., and T. A. Steitz. 2000. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10:117–123. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
