Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1)
- PMID: 11784859
- PMCID: PMC133546
- DOI: 10.1128/MCB.22.3.835-848.2002
Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1)
Abstract
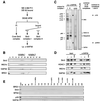
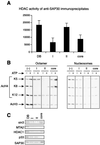
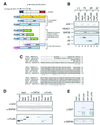
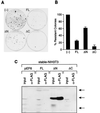
Sin3 is an evolutionarily conserved corepressor that exists in different complexes with the histone deacetylases HDAC1 and HDAC2. Sin3-HDAC complexes are believed to deacetylate nucleosomes in the vicinity of Sin3-regulated promoters, resulting in a repressed chromatin structure. We have previously found that a human Sin3-HDAC complex includes HDAC1 and HDAC2, the histone-binding proteins RbAp46 and RbAp48, and two novel polypeptides SAP30 and SAP18. SAP30 is a specific component of Sin3 complexes since it is absent in other HDAC1/2-containing complexes such as NuRD. SAP30 mediates interactions with different polypeptides providing specificity to Sin3 complexes. We have identified p33ING1b, a negative growth regulator involved in the p53 pathway, as a SAP30-associated protein. Two distinct Sin3-p33ING1b-containing complexes were isolated, one of which associates with the subunits of the Brg1-based Swi/Snf chromatin remodeling complex. The N terminus of p33ING1b, which is divergent among a family of ING1 polypeptides, associates with the Sin3 complex through direct interaction with SAP30. The N-terminal domain of p33 is present in several uncharacterized human proteins. We show that overexpression of p33ING1b suppresses cell growth in a manner dependent on the intact Sin3-HDAC-interacting domain.
Figures


References
-
- Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePlnho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49–55. - PubMed
-
- Ausio, J., and K. E. van Holde. 1986. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry 25:1421–1428. - PubMed
-
- Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by temary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767–776. - PubMed
-
- Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
