The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation
- PMID: 11784867
- PMCID: PMC133540
- DOI: 10.1128/MCB.22.3.927-934.2002
The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation
Abstract
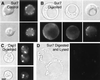
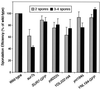
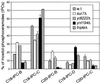
We have discovered a novel cortical patch structure in Saccharomyces cerevisiae defined by a family of integral plasma membrane proteins, including Sur7p, Ynl194p, and Ydl222p. Sur7p-family patches localized as cortical patches that were immobile and stable. These patches were polarized to regions of the cell with a mature cell wall; they were absent from small buds and the tips of many medium-sized buds. These patches were distinct from other known cortical structures. Digestion of the cell wall caused Sur7p patches to disassemble, indicating that Sur7p requires cell wall-dependent extracellular interactions for its localization as patches. sur7Delta, ydl222Delta, and ynl194Delta mutants had reduced sporulation efficiencies. SUR7 was originally described as a multicopy suppressor of rvs167, whose product is an actin patch component. This suppression is probably mediated by sphingolipids, since deletion of SUR7, YDL222, and YNL194 altered the sphingolipid content of the yeast plasma membrane, and other SUR genes suppress rvs167 via effects on sphingolipid synthesis. In particular, the sphingoid base length and number of hydroxyl groups in inositol phosphorylceramides were altered in sur7Delta, ydl222Delta, and yne194Delta strains.
Figures




References
-
- Balguerie, A., P. Sivadon, M. Bonneu, and M. Aigle. 1999. Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112:2529–2537. - PubMed
-
- Barral, Y., V. Mermall, M. S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5:841–851. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
