Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival
- PMID: 11784871
- PMCID: PMC133541
- DOI: 10.1128/MCB.22.3.965-977.2002
Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival
Abstract
Class Ia phosphoinositide (PI) 3-kinase is a central component in growth factor signaling and is comprised of a p110 catalytic subunit and a regulatory subunit, the most common family of which is derived from the p85alpha gene (Pik3r1). Optimal signaling through the PI 3-kinase pathway depends on a critical molecular balance between the regulatory and catalytic subunits. In wild-type cells, the p85 subunit is more abundant than p110, leading to competition between the p85 monomer and the p85-p110 dimer and ineffective signaling. Heterozygous disruption of Pik3r1 results in increased Akt activity and decreased apoptosis by insulin-like growth factor 1 (IGF-1) through up-regulated phosphatidylinositol (3,4,5)-triphosphate production. Complete depletion of p85alpha, on the other hand, results in significantly increased apoptosis due to reduced PI 3-kinase-dependent signaling. Thus, a reduction in p85alpha represents a novel therapeutic target for enhancing IGF-1/insulin signaling, prolongation of cell survival, and protection against apoptosis.
Figures


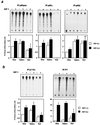



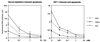

Similar articles
-
Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling.J Biol Chem. 2003 Nov 28;278(48):48453-66. doi: 10.1074/jbc.M305602200. Epub 2003 Sep 22. J Biol Chem. 2003. Retraction in: J Biol Chem. 2017 Mar 31;292(13):5608. doi: 10.1074/jbc.A117.305602. PMID: 14504291 Retracted.
-
Reduced expression of the murine p85alpha subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes.J Clin Invest. 2002 Jan;109(1):141-9. doi: 10.1172/JCI13305. J Clin Invest. 2002. PMID: 11781359 Free PMC article.
-
The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex.J Cell Biol. 2005 Aug 1;170(3):455-64. doi: 10.1083/jcb.200503088. Epub 2005 Jul 25. J Cell Biol. 2005. PMID: 16043515 Free PMC article.
-
Class IA PI3K regulatory subunits: p110-independent roles and structures.Biochem Soc Trans. 2020 Aug 28;48(4):1397-1417. doi: 10.1042/BST20190845. Biochem Soc Trans. 2020. PMID: 32677674 Free PMC article. Review.
-
Molecular Mechanisms of Human Disease Mediated by Oncogenic and Primary Immunodeficiency Mutations in Class IA Phosphoinositide 3-Kinases.Front Immunol. 2018 Mar 19;9:575. doi: 10.3389/fimmu.2018.00575. eCollection 2018. Front Immunol. 2018. PMID: 29616047 Free PMC article. Review.
Cited by
-
PI3K at the crossroads of tumor angiogenesis signaling pathways.Mol Cell Oncol. 2015 Feb 26;2(2):e975624. doi: 10.4161/23723556.2014.975624. eCollection 2015 Apr-Jun. Mol Cell Oncol. 2015. PMID: 27308431 Free PMC article. Review.
-
Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth.J Clin Invest. 2006 Jan;116(1):101-14. doi: 10.1172/JCI25735. Epub 2005 Dec 22. J Clin Invest. 2006. PMID: 16374520 Free PMC article.
-
The regulatory subunits of PI3K, p85α and p85β, differentially affect BRD7-mediated regulation of insulin signaling.J Mol Cell Biol. 2022 Jan 29;13(12):889-901. doi: 10.1093/jmcb/mjab073. J Mol Cell Biol. 2022. PMID: 34751372 Free PMC article.
-
Acute effects of leptin require PI3K signaling in hypothalamic proopiomelanocortin neurons in mice.J Clin Invest. 2008 May;118(5):1796-805. doi: 10.1172/JCI32964. J Clin Invest. 2008. PMID: 18382766 Free PMC article.
-
Suppressor of cytokine signaling (SOCS)5 ameliorates influenza infection via inhibition of EGFR signaling.Elife. 2017 Feb 14;6:e20444. doi: 10.7554/eLife.20444. Elife. 2017. PMID: 28195529 Free PMC article.
References
-
- Baltensperger, K., L. M. Kozma, S. R. Jaspers, and M. P. Czech. 1994. Regulation by insulin of phosphatidylinositol 3′-kinase bound to alpha- and beta-isoforms of p85 regulatory subunit. J. Biol. Chem. 269:28937–28946. - PubMed
-
- Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358–1362. - PubMed
-
- Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857–868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
