Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases
- PMID: 11788710
- PMCID: PMC99834
- DOI: 10.1093/nar/30.2.475
Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases
Abstract
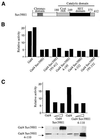
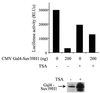
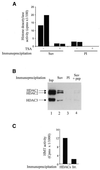
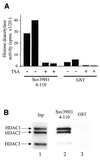
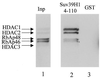
The histone methyl transferase Suv39H1 is involved in silencing by pericentric heterochromatin. It specifically methylates K9 of histone H3, thereby creating a high affinity binding site for HP1 proteins. We and others have shown recently that it is also involved in transcriptional repression by the retinoblastoma protein Rb. Strikingly, both HP1 localisation and repression by Rb also require, at least in part, histone deacetylases. We found here that repression of a heterologous promoter by Suv39H1 is dependent on histone deacetylase activity. However, the enzymatic activity of Suv39H1 is not required, since the N-terminal part is by itself a transcriptional repression domain. Coimmunoprecipitation experiments indicated that Suv39H1 can physically interact with HDAC1, -2 and -3, therefore suggesting that transcriptional repression by Suv39H1 could be the consequence of histone deacetylases recruitment. Consistent with this interpretation, the N-terminal transcriptional repression domain of Suv39H1 bound the so-called 'core histone deacetylase complex', composed of HDAC1, HDAC2 and the Rb-associated proteins RbAp48 and RbAp46. Taken together, our results suggest that a complex containing both the Suv39H1 histone methyl transferase and histone deacetylases could be involved in heterochromatin silencing or transcriptional repression by Rb.
Figures
References
-
- Wade P.A., Pruss,D. and Wolffe,A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. - PubMed
-
- Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. - PubMed
-
- Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. - PubMed
-
- Grewal S.I. (2000) Transcriptional silencing in fission yeast. J. Cell Physiol., 184, 311–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

