PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors
- PMID: 11788728
- PMCID: PMC99841
- DOI: 10.1093/nar/30.2.e2
PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors
Abstract
We introduce a PCR-based procedure for generating a gene disruption construct. This method depends on DNA fragment fusion by the PCR technique and requires only two steps of PCR to obtain a sufficient amount of the gene disruption construct for one transformation experiment. The first step involves three separate PCR syntheses of a selectable marker cassette and the 5'- and 3'-regions of a target gene. Of the four primers used in amplification of the 5'- and 3'-regions of the target gene, two primers placed proximal to the site of the marker cassette are designed to have sequence tags complementary to the 5'- or 3'-side of the marker cassette. The two primers used in PCR synthesis of the marker cassette are complementary to the tagged primers. By fusion PCR, the 5' and 3' PCR products are linked to the marker cassette via the regions of tagged primers that overlap. A sufficient amount of the disruption construct can be directly amplified with the outermost primers. This method is simple, rapid and relatively inexpensive. In addition, there is the freedom of attaching long flanking regions to any selectable marker cassette.
Figures
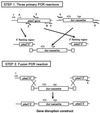
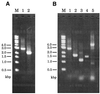


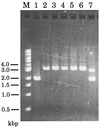
References
-
- Kessin R. (2001) Dictyostelium; Evolution, Cell Biology and the Development of Multicellularity. Cambridge University Press, Cambridge, UK.
-
- De Lozanne A. and Spudich,J.A. (1987) Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science, 236, 1086–1091. - PubMed
-
- Mann S.K. and Firtel,R.A. (1991) A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech. Dev., 35, 89–101. - PubMed
-
- Harwood A., Plyte,S.E., Woodgett,J., Strutt,H. and Kay,R.R. (1995) Glycogen synthetase kinase 3 regulates cell fate in Dictyostelium. Cell, 80, 139–148. - PubMed
-
- Morio T., Urushihara,H., Saito,T., Ugawa,Y., Mizuno,H., Yoshida,M., Yoshino,R., Mitra,B.N., Pi,M., Sato,T., Takemoto,K., Yasukawa,H., Williams,J., Maeda,M., Takeuchi,I., Ochiai,H. and Tanaka,Y. (1998) The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res., 31, 335–340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

