A new approach to genome mapping and sequencing: slalom libraries
- PMID: 11788732
- PMCID: PMC99845
- DOI: 10.1093/nar/30.2.e6
A new approach to genome mapping and sequencing: slalom libraries
Abstract
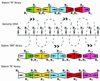
We describe here an efficient strategy for simultaneous genome mapping and sequencing. The approach is based on physically oriented, overlapping restriction fragment libraries called slalom libraries. Slalom libraries combine features of general genomic, jumping and linking libraries. Slalom libraries can be adapted to different applications and two main types of slalom libraries are described in detail. This approach was used to map and sequence (with approximately 46% coverage) two human P1-derived artificial chromosome (PAC) clones, each of approximately 100 kb. This model experiment demonstrates the feasibility of the approach and shows that the efficiency (cost-effectiveness and speed) of existing mapping/sequencing methods could be improved at least 5-10-fold. Furthermore, since the efficiency of contig assembly in the slalom approach is virtually independent of length of sequence reads, even short sequences produced by rapid, high throughput sequencing techniques would suffice to complete a physical map and a sequence scan of a small genome.
Figures
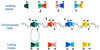




References
-
- Dunham I., Hunt,A.R., Collins,J.E., Bruskiewich,R., Beare,D.M., Clamp,M., Smink,L.J., Ainscough,R., Almeida,J.P., Babbage,A. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. - PubMed
-
- Hattory M., Fujiyama,A., Taylor,T.D., Watanabe,H., Yada,T., Park,H.-S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.-K. et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. - PubMed
-
- Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. - PubMed
-
- Myers E.W., Sutton,G.G., Delcher,A.L., Dew,I.M., Fasulo,D.P., Flanigan,M.J., Kravitz,S.A., Mobarry,C.M., Reinert,K.H., Remington,K.A. et al. (2000) A whole-genome assembly of Drosophila. Science, 287, 2196–2204. - PubMed
-
- Broder S. and Venter,J.C. (2000) Sequencing the entire genomes of free-living organisms: the foundation of pharmacology in the new millennium. Annu. Rev. Pharmacol. Toxicol., 40, 97–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

