AtFXG1, an Arabidopsis gene encoding alpha-L-fucosidase active against fucosylated xyloglucan oligosaccharides
- PMID: 11788770
- PMCID: PMC148987
AtFXG1, an Arabidopsis gene encoding alpha-L-fucosidase active against fucosylated xyloglucan oligosaccharides
Abstract
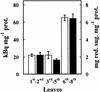
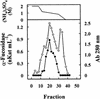
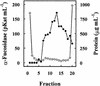
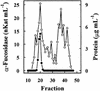
An alpha-L-fucosidase (EC 3.2.1.51) able to release the t-fucosyl residue from the side chain of xyloglucan oligosaccharides has been detected in the leaves of Arabidopsis plants. Moreover, an alpha-L-fucosidase with similar substrate specificity was purified from cabbage (Brassica oleracea) leaves to render a single band on SDS-PAGE. Two peptide sequences were obtained from this protein band, and they were used to identify an Arabidopsis gene coding for an alpha-fucosidase that we propose to call AtFXG1. In addition, an Arabidopsis gene with homology with known alpha-L-fucosidases has been also found, and we proposed to name it as AtFUC1. Both AtFXG1 and ATFUC1 were heterologously expressed in Pichia pastoris cells and the alpha-L-fucosidase activities secreted to the culture medium. The alpha-L-fucosidase encoded by AtFXG1 was active against the oligosaccharides from xyloglucan XXFG as well as against 2'-fucosyl-lactitol but not against p-nitrophenyl-alpha-L-fucopyranoside. However, the AtFUC1 heterologously expressed was active only against 2'-fucosyl-lactitol. Thus, the former must be related to xyloglucan metabolism.
Figures



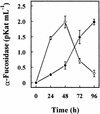
Similar articles
-
Identification of an Arabidopsis gene encoding a GH95 alpha1,2-fucosidase active on xyloglucan oligo- and polysaccharides.Phytochemistry. 2008 Jul;69(10):1983-8. doi: 10.1016/j.phytochem.2008.03.024. Epub 2008 May 19. Phytochemistry. 2008. PMID: 18495185
-
Cloning and expression pattern of a gene encoding an alpha-xylosidase active against xyloglucan oligosaccharides from Arabidopsis.Plant Physiol. 2001 Jun;126(2):910-20. doi: 10.1104/pp.126.2.910. Plant Physiol. 2001. PMID: 11402218 Free PMC article.
-
Purification, characterization, and cell wall localization of an alpha-fucosidase that inactivates a xyloglucan oligosaccharin.Plant J. 1993 Mar;3(3):415-26. doi: 10.1046/j.1365-313x.1993.t01-24-00999.x. Plant J. 1993. PMID: 8220450
-
α-L-Fucosidases and their applications for the production of fucosylated human milk oligosaccharides.Appl Microbiol Biotechnol. 2020 Jul;104(13):5619-5631. doi: 10.1007/s00253-020-10635-7. Epub 2020 May 1. Appl Microbiol Biotechnol. 2020. PMID: 32356197 Review.
-
Structure and function of microbial α-l-fucosidases: a mini review.Essays Biochem. 2023 Apr 18;67(3):399-414. doi: 10.1042/EBC20220158. Essays Biochem. 2023. PMID: 36805644 Free PMC article. Review.
Cited by
-
A composite transcriptional signature differentiates responses towards closely related herbicides in Arabidopsis thaliana and Brassica napus.Plant Mol Biol. 2010 Mar;72(4-5):545-56. doi: 10.1007/s11103-009-9590-y. Epub 2009 Dec 31. Plant Mol Biol. 2010. PMID: 20043233 Free PMC article.
-
AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides.Plant Cell. 2011 Nov;23(11):4025-40. doi: 10.1105/tpc.111.089193. Epub 2011 Nov 11. Plant Cell. 2011. PMID: 22080600 Free PMC article.
-
Evolution of xyloglucan-related genes in green plants.BMC Evol Biol. 2010 Nov 5;10:341. doi: 10.1186/1471-2148-10-341. BMC Evol Biol. 2010. PMID: 21054875 Free PMC article.
-
The fuc1 gene product (20 kDa FUC1) of Pisum sativum has no alpha-L-fucosidase activity.Plant Mol Biol. 2003 Apr;51(6):877-84. doi: 10.1023/a:1023053101938. Plant Mol Biol. 2003. PMID: 12777048
-
Origins and Evolution of the α-L-Fucosidases: From Bacteria to Metazoans.Front Microbiol. 2019 Aug 27;10:1756. doi: 10.3389/fmicb.2019.01756. eCollection 2019. Front Microbiol. 2019. PMID: 31507539 Free PMC article.
References
-
- Augur C, Benhamou N, Darvill A, Albersheim P. Purification, characterization, and cell wall localization of an α-fucosidase that inactivates a xyloglucan oligosaccharin. Plant J. 1993;3:415–426. - PubMed
-
- Augur C, Stiefel V, Darvill A, Albersheim P, Puigdomenech P. Molecular cloning and pattern of expression of an α-l-fucosidase gene from pea seedlings. J Biol Chem. 1995;270:24839–24843. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Brick DJ, Brumlik MJ, Buckley T, Cao JX, Davies PC, Misra S, Tranbarger TJ, Upton C. A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett. 1995;377:475–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
