Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity
- PMID: 11790734
- PMCID: PMC139515
- DOI: 10.1128/JB.184.3.654-665.2002
Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity
Abstract
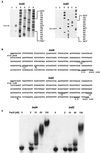
In the plant-pathogenic bacterium Erwinia chrysanthemi production of pectate lyases, the main virulence determinant, is modulated by a complex network involving several regulatory proteins. One of these regulators, PecS, also controls the synthesis of a blue pigment identified as indigoidine. Since production of this pigment is cryptic in the wild-type strain, E. chrysanthemi ind mutants deficient in indigoidine synthesis were isolated by screening a library of Tn5-B21 insertions in a pecS mutant. These ind mutations were localized close to the regulatory pecS-pecM locus, immediately downstream of pecM. Sequence analysis of this DNA region revealed three open reading frames, indA, indB, and indC, involved in indigoidine biosynthesis. No specific function could be assigned to IndA. In contrast, IndB displays similarity to various phosphatases involved in antibiotic synthesis and IndC reveals significant homology with many nonribosomal peptide synthetases (NRPS). The IndC product contains an adenylation domain showing the signature sequence DAWCFGLI for glutamine recognition and an oxidation domain similar to that found in various thiazole-forming NRPS. These data suggest that glutamine is the precursor of indigoidine. We assume that indigoidine results from the condensation of two glutamine molecules that have been previously cyclized by intramolecular amide bond formation and then dehydrogenated. Expression of ind genes is strongly derepressed in the pecS background, indicating that PecS is the main regulator of this secondary metabolite synthesis. DNA band shift assays support a model whereby the PecS protein represses indA and indC expression by binding to indA and indC promoter regions. The regulatory link, via pecS, between indigoidine and virulence factor production led us to explore a potential role of indigoidine in E. chrysanthemi pathogenicity. Mutants impaired in indigoidine production were unable to cause systemic invasion of potted Saintpaulia ionantha. Moreover, indigoidine production conferred an increased resistance to oxidative stress, indicating that indigoidine may protect the bacteria against the reactive oxygen species generated during the plant defense response.
Figures






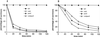

References
-
- Altschul, A. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
-
- Bardonnet, N., and C. Blanco. 1992. uidA antibiotic resistance cassettes for insertion mutagenesis, gene fusion and genetic constructions. FEMS Microbiol. Lett. 93:243–248. - PubMed
-
- Boccara, M., S. Tandon, and A. d’Harlingue. 1994. Studies of Erwinia chrysanthemi interactions with plant tissue culture cells, abstr. 176, p.58. In Proceedings of the Seventh International Symposium on Molecular Plant-Microbe Interactions, Edinburgh, Scotland, 26 June to 1 July 1994.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

