Nested evolution of a tRNA(Leu)(UAA) group I intron by both horizontal intron transfer and recombination of the entire tRNA locus
- PMID: 11790735
- PMCID: PMC139512
- DOI: 10.1128/JB.184.3.666-671.2002
Nested evolution of a tRNA(Leu)(UAA) group I intron by both horizontal intron transfer and recombination of the entire tRNA locus
Abstract
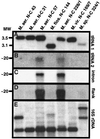
The origin and evolution of bacterial introns are still controversial issues. Here we present data on the distribution and evolution of a recently discovered divergent tRNA(Leu)(UAA) intron. The intron shows a higher sequence affiliation with introns in tRNA(Ile)(CAU) and tRNA(Arg)(CCU) genes in alpha- and beta-proteobacteria, respectively, than with other cyanobacterial tRNA(Leu)(UAA) group I introns. The divergent tRNA(Leu)(UAA) intron is sporadically distributed both within the Nostoc and the Microcystis radiations. The complete tRNA gene, including flanking regions and intron from Microcystis aeruginosa strain NIVA-CYA 57, was sequenced in order to elucidate the evolutionary pattern of this intron. Phylogenetic reconstruction gave statistical evidence for different phylogenies for the intron and exon sequences, supporting an evolutionary model involving horizontal intron transfer. The distribution of the tRNA gene, its flanking regions, and the introns were addressed by Southern hybridization and PCR amplification. The tRNA gene, including the flanking regions, were absent in the intronless stains but present in the intron-containing strains. This suggests that the sporadic distribution of this intron within the Microcystis genus cannot be attributed to intron mobility but rather to an instability of the entire tRNA(Leu)(UAA) intron-containing genome region. Taken together, the complete data set for the evolution of this intron can best be explained by a model involving a nested evolution of the intron, i.e., wherein the intron has been transferred horizontally (probably through a single or a few events) to a tRNA(Leu)(UAA) gene which is located within a unstable genome region.
Figures
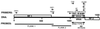
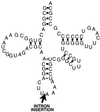




References
-
- Besendahl, A., Y. L. Qiu, J. Lee, J. D. Palmer, and D. Bhattacharya. 2000. The cyanobacterial origin and vertical transmission of the plastid tRNA(Leu) group-I intron. Curr. Genet. 37:12–23. - PubMed
-
- Bonocora, R. P., and D. A. Shub. 2001. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA fMet genes. Mol. Microbiol. 39:1299–1306. - PubMed
-
- Castenholz, R. W., and J. B. Waterbury. 1989. Group I. Cyanobacteria, p.1710–1728. In N. Pfenning and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

