Integration of global regulation of two aromatic-responsive sigma(54)-dependent systems: a common phenotype by different mechanisms
- PMID: 11790746
- PMCID: PMC139538
- DOI: 10.1128/JB.184.3.760-770.2002
Integration of global regulation of two aromatic-responsive sigma(54)-dependent systems: a common phenotype by different mechanisms
Abstract
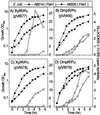
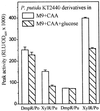
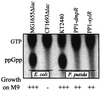
Pseudomonas-derived regulators DmpR and XylR are structurally and mechanistically related sigma(54)-dependent activators that control transcription of genes involved in catabolism of aromatic compounds. The binding of distinct sets of aromatic effectors to these regulatory proteins results in release of a repressive interdomain interaction and consequently allows the activators to promote transcription from their cognate target promoters. The DmpR-controlled Po promoter region and the XylR-controlled Pu promoter region are also similar, although homology is limited to three discrete DNA signatures for binding sigma(54) RNA polymerase, the integration host factor, and the regulator. These common properties allow cross-regulation of Pu and Po by DmpR and XylR in response to appropriate aromatic effectors. In vivo, transcription of both the DmpR/Po and XylR/Pu regulatory circuits is subject to dominant global regulation, which results in repression of transcription during growth in rich media. Here, we comparatively assess the contribution of (p)ppGpp, the FtsH protease, and a component of an alternative phosphoenolpyruvate-sugar phosphotransferase system, which have been independently implicated in mediating this level of regulation. Further, by exploiting the cross-regulatory abilities of these two circuits, we identify the target component(s) that are intercepted in each case. The results show that (i) contrary to previous speculation, FtsH is not universally required for transcription of sigma(54)-dependent systems; (ii) the two factors found to impact the XylR/Pu regulatory circuit do not intercept the DmpR/Po circuit; and (iii) (p)ppGpp impacts the DmpR/Po system to a greater extent than the XylR/Pu system in both the native Pseudomonas putida and a heterologous Escherichia coli host. The data demonstrate that, despite the similarities of the specific regulatory circuits, the host global regulatory network latches onto and dominates over these specific circuits by exploiting their different properties. The mechanistic implications of how each of the host factors exerts its action are discussed.
Figures



References
-
- Abril, M. A., M. Buck, and J. L. Ramos. 1991. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J. Biol. Chem. 266:15832–15838. - PubMed
-
- Aviv, M., H. Giladi, G. Schreiber, A. B. Oppenheim, and G. Glaser. 1994. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase. rpoS, ppGpp and by autoregulation. Mol. Microbiol. 14:1021–1031. - PubMed
-
- Blaszczak, A., C. Georgopoulos, and K. Liberek. 1999. On the mechanism of FtsH-dependent degradation of the ς32 transcriptional regulator of Escherichia coli and the role of the DnaK chaperone machine. Mol. Microbiol. 31:157–166. - PubMed
-
- Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759–9769. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

