Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products
- PMID: 11790749
- PMCID: PMC139519
- DOI: 10.1128/JB.184.3.785-793.2002
Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products
Abstract
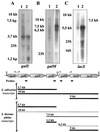
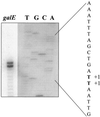
Streptococcus salivarius is a lactose- and galactose-positive bacterium that is phylogenetically closely related to Streptococcus thermophilus, a bacterium that metabolizes lactose but not galactose. In this paper, we report a comparative characterization of the S. salivarius and S. thermophilus gal-lac gene clusters. The clusters have the same organization with the order galR (codes for a transcriptional regulator and is transcribed in the opposite direction), galK (galactokinase), galT (galactose-1-P uridylyltransferase), galE (UDP-glucose 4-epimerase), galM (galactose mutarotase), lacS (lactose transporter), and lacZ (beta-galactosidase). An analysis of the nucleotide sequence as well as Northern blotting and primer extension experiments revealed the presence of four promoters located upstream from galR, the gal operon, galM, and the lac operon of S. salivarius. Putative promoters with virtually identical nucleotide sequences were found at the same positions in the S. thermophilus gal-lac gene cluster. An additional putative internal promoter at the 3' end of galT was found in S. thermophilus but not in S. salivarius. The results clearly indicated that the gal-lac gene cluster was efficiently transcribed in both species. The Shine-Dalgarno sequences of galT and galE were identical in both species, whereas the ribosome binding site of S. thermophilus galK differed from that of S. salivarius by two nucleotides, suggesting that the S. thermophilus galK gene might be poorly translated. This was confirmed by measurements of enzyme activities.
Figures





Similar articles
-
Characterization of a galactokinase-positive recombinant strain of Streptococcus thermophilus.Appl Environ Microbiol. 2004 Aug;70(8):4596-603. doi: 10.1128/AEM.70.8.4596-4603.2004. Appl Environ Microbiol. 2004. PMID: 15294791 Free PMC article.
-
Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase.J Bacteriol. 1990 Jul;172(7):4037-47. doi: 10.1128/jb.172.7.4037-4047.1990. J Bacteriol. 1990. PMID: 1694527 Free PMC article.
-
Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus.J Bacteriol. 2001 Feb;183(4):1184-94. doi: 10.1128/JB.183.4.1184-1194.2001. J Bacteriol. 2001. PMID: 11157930 Free PMC article.
-
Hereditary galactosemia.Metabolism. 2018 Jun;83:188-196. doi: 10.1016/j.metabol.2018.01.025. Epub 2018 Jan 31. Metabolism. 2018. PMID: 29409891 Review.
-
Molecular basis of disorders of human galactose metabolism: past, present, and future.Mol Genet Metab. 2000 Sep-Oct;71(1-2):62-5. doi: 10.1006/mgme.2000.3073. Mol Genet Metab. 2000. PMID: 11001796 Review.
Cited by
-
Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway.J Bacteriol. 2003 Feb;185(3):870-8. doi: 10.1128/JB.185.3.870-878.2003. J Bacteriol. 2003. PMID: 12533462 Free PMC article.
-
Characterization of a galactokinase-positive recombinant strain of Streptococcus thermophilus.Appl Environ Microbiol. 2004 Aug;70(8):4596-603. doi: 10.1128/AEM.70.8.4596-4603.2004. Appl Environ Microbiol. 2004. PMID: 15294791 Free PMC article.
-
Role of galK and galM in galactose metabolism by Streptococcus thermophilus.Appl Environ Microbiol. 2008 Feb;74(4):1264-7. doi: 10.1128/AEM.01585-07. Epub 2007 Dec 7. Appl Environ Microbiol. 2008. PMID: 18065633 Free PMC article.
-
Characterization of genes involved in the metabolism of alpha-galactosides by Lactococcus raffinolactis.Appl Environ Microbiol. 2003 Jul;69(7):4049-56. doi: 10.1128/AEM.69.7.4049-4056.2003. Appl Environ Microbiol. 2003. PMID: 12839781 Free PMC article.
-
Milk fermentation by monocultures or co-cultures of Streptococcus thermophilus strains.Front Bioeng Biotechnol. 2022 Dec 12;10:1097013. doi: 10.3389/fbioe.2022.1097013. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36578511 Free PMC article.
References
-
- Ajdic, D., I. Sutcliffe, R. R. B. Russell, and J. J. Ferretti. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137–144. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
-
- Avigad, G., D. Amaval, C. Ascension, and B. L. Horecker. 1962. The D-galactose oxydase of Polyporus circinatus. J. Biol. Chem. 237:2736–2743. - PubMed
-
- de Vos, W. M., and E. E. Vaughan. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217–237. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

