The structure of spider toxin huwentoxin-II with unique disulfide linkage: evidence for structural evolution
- PMID: 11790834
- PMCID: PMC2373433
- DOI: 10.1110/ps.30502
The structure of spider toxin huwentoxin-II with unique disulfide linkage: evidence for structural evolution
Abstract
The three-dimensional structure of huwentoxin-II (HWTX-II), an insecticidal peptide purified from the venom of spider Selenocosmia huwena with a unique disulfide bond linkage as I-III, II-V, and IV-VI, has been determined using 2D (1)H-NMR. The resulting structure of HWTX-II contains two beta-turns (C4-S7 and K24-W27) and a double-stranded antiparallel beta-sheet (W27-C29 and C34-K36). Although the C-terminal double-stranded beta-sheet cross-linked by two disulfide bonds (II-V and IV-VI in HWTX-II, II-V and III-VI in the ICK molecules) is conserved both in HWTX-II and the ICK molecules, the structure of HWTX-II is unexpected absence of the cystine knot because of its unique disulfide linkage. It suggests that HWTX-II adopts a novel scaffold different from the ICK motif that is adopted by all other spider toxin structures elucidated thus far. Furthermore, the structure of HWTX-II, which conforms to the disulfide-directed beta-hairpin (DDH) motif, not only supports the hypothesis that the ICK is a minor elaboration of the more ancestral DDH motif but also suggests that HWTX-II may have evolved from the same structural ancestor.
Figures

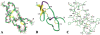
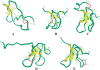
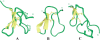
References
-
- Anette, S.N. 1989. Tarantula (Eurypelma californicum) venom, a multicomponent system. Biol. Chem. Hoppe-Seyler. 370: 485–498. - PubMed
-
- Brünger, A.T. 1992. X-PLOR manual, Version 3.1. Yale University, New Haven, CT.
-
- Fletcher, J.I., Smith, R., O'Donoghue, S.I., Nilges, M., Connor, M., Howden, M.E., Christie, M.J., and King, G.F. 1997a. The structure of a novel insecticidal neurotoxin, ω-atracotoxin-HV1, from the venom of an Australian funnel web spider. Nat. Struct. Biol. 4:559–566. - PubMed
-
- Fletcher, J.I., Chapman, B.E., Mackay, J.P., Howden, M.E., and King, G.F. 1997b. The structure of versutoxin (δ-atracotoxin-Hv1) provides insights into the binding of site 3 neurotoxins to the voltage-gated sodium channel. Structure (London). 5: 1525–1535. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

