Structural studies of the pigeon cytosolic NADP(+)-dependent malic enzyme
- PMID: 11790843
- PMCID: PMC2373443
- DOI: 10.1110/ps.38002
Structural studies of the pigeon cytosolic NADP(+)-dependent malic enzyme
Abstract
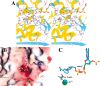
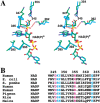
Malic enzymes are widely distributed in nature, and have important biological functions. They catalyze the oxidative decarboxylation of malate to produce pyruvate and CO(2) in the presence of divalent cations (Mg(2+), Mn(2+)). Most malic enzymes have a clear selectivity for the dinucleotide cofactor, being able to use either NAD(+) or NADP(+), but not both. Structural studies of the human mitochondrial NAD(+)-dependent malic enzyme established that malic enzymes belong to a new class of oxidative decarboxylases. Here we report the crystal structure of the pigeon cytosolic NADP(+)-dependent malic enzyme, in a closed form, in a quaternary complex with NADP(+), Mn(2+), and oxalate. This represents the first structural information on an NADP(+)-dependent malic enzyme. Despite the sequence conservation, there are large differences in several regions of the pigeon enzyme structure compared to the human enzyme. One region of such differences is at the binding site for the 2'-phosphate group of the NADP(+) cofactor, which helps define the cofactor selectivity of the enzymes. Specifically, the structural information suggests Lys362 may have an important role in the NADP(+) selectivity of the pigeon enzyme, confirming our earlier kinetic observations on the K362A mutant. Our structural studies also revealed differences in the organization of the tetramer between the pigeon and the human enzymes, although the pigeon enzyme still obeys 222 symmetry.
Figures


Similar articles
-
Potent and competitive inhibition of malic enzymes by lanthanide ions.Biochem Biophys Res Commun. 2000 Aug 2;274(2):440-4. doi: 10.1006/bbrc.2000.3163. Biochem Biophys Res Commun. 2000. PMID: 10913357
-
Cloning and expression of pigeon liver cytosolic NADP(+)-dependent malic enzyme cDNA and some of its abortive mutants.Arch Biochem Biophys. 1994 Apr;310(1):158-66. doi: 10.1006/abbi.1994.1152. Arch Biochem Biophys. 1994. PMID: 8161199
-
Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme: a new class of oxidative decarboxylases.Structure. 1999 Aug 15;7(8):R877-89. Structure. 1999. PMID: 10477256
-
NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways.FEBS Lett. 2001 Feb 9;490(1-2):1-6. doi: 10.1016/s0014-5793(00)02331-0. FEBS Lett. 2001. PMID: 11172800 Review.
-
NADP-malic enzyme from plants.Phytochemistry. 1992 Jun;31(6):1845-57. doi: 10.1016/0031-9422(92)80322-6. Phytochemistry. 1992. PMID: 1368216 Review.
Cited by
-
A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis.Plant Physiol. 2005 Sep;139(1):39-51. doi: 10.1104/pp.105.065953. Epub 2005 Aug 19. Plant Physiol. 2005. PMID: 16113210 Free PMC article.
-
Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum.Foods. 2022 Apr 1;11(7):1031. doi: 10.3390/foods11071031. Foods. 2022. PMID: 35407118 Free PMC article.
-
Characterization of an archaeal malic enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1.Archaea. 2005 May;1(5):293-301. doi: 10.1155/2005/250757. Archaea. 2005. PMID: 15876562 Free PMC article.
-
Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human mitochondrial NAD(P)+-dependent malate dehydrogenase (malic enzyme).Biochem J. 2005 Nov 15;392(Pt 1):39-45. doi: 10.1042/BJ20050641. Biochem J. 2005. PMID: 15989682 Free PMC article.
-
Comparative Approach of the de novo Fatty Acid Synthesis (Lipogenesis) between Ruminant and Non Ruminant Mammalian Species: From Biochemical Level to the Main Regulatory Lipogenic Genes.Curr Genomics. 2010 May;11(3):168-83. doi: 10.2174/138920210791110960. Curr Genomics. 2010. PMID: 21037855 Free PMC article.
References
-
- Bhargava, G., Mui, S., Pav, S., Wu, H., Loeber, G., and Tong, L. 1999. Preliminary crystallographic studies of human mitochondrial NAD(P)+-dependent malic enzyme. J. Struct. Biol. 127: 72–75. - PubMed
-
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., Read, R.J., Rice, L.M., Simonson, T., and Warren, G.L. 1998. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. - PubMed
-
- Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graphics 5: 103–106.
-
- CCP4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50: 760–763. - PubMed
-
- Chang, G.-G. and Hsu, R.Y. 1977. Mechanism of pigeon liver malic enzyme. Kinetics, specificity, and half-site stoichiometry of the alkylation of a cysteinyl residue by the substrate-inhibitor bromopyruvate. Biochemistry 16: 311–320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

