A genomewide scan identifies two novel loci involved in specific language impairment
- PMID: 11791209
- PMCID: PMC384915
- DOI: 10.1086/338649
A genomewide scan identifies two novel loci involved in specific language impairment
Abstract
Approximately 4% of English-speaking children are affected by specific language impairment (SLI), a disorder in the development of language skills despite adequate opportunity and normal intelligence. Several studies have indicated the importance of genetic factors in SLI; a positive family history confers an increased risk of development, and concordance in monozygotic twins consistently exceeds that in dizygotic twins. However, like many behavioral traits, SLI is assumed to be genetically complex, with several loci contributing to the overall risk. We have compiled 98 families drawn from epidemiological and clinical populations, all with probands whose standard language scores fall > or =1.5 SD below the mean for their age. Systematic genomewide quantitative-trait-locus analysis of three language-related measures (i.e., the Clinical Evaluation of Language Fundamentals-Revised [CELF-R] receptive and expressive scales and the nonword repetition [NWR] test) yielded two regions, one on chromosome 16 and one on 19, that both had maximum LOD scores of 3.55. Simulations suggest that, of these two multipoint results, the NWR linkage to chromosome 16q is the most significant, with empirical P values reaching 10(-5), under both Haseman-Elston (HE) analysis (LOD score 3.55; P=.00003) and variance-components (VC) analysis (LOD score 2.57; P=.00008). Single-point analyses provided further support for involvement of this locus, with three markers, under the peak of linkage, yielding LOD scores >1.9. The 19q locus was linked to the CELF-R expressive-language score and exceeds the threshold for suggestive linkage under all types of analysis performed-multipoint HE analysis (LOD score 3.55; empirical P=.00004) and VC (LOD score 2.84; empirical P=.00027) and single-point HE analysis (LOD score 2.49) and VC (LOD score 2.22). Furthermore, both the clinical and epidemiological samples showed independent evidence of linkage on both chromosome 16q and chromosome 19q, indicating that these may represent universally important loci in SLI and, thus, general risk factors for language impairment.
Figures
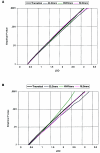

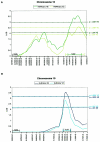
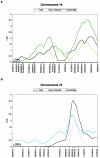
References
Electronic-Database Information
-
- Center for Statistical Genetics, University of Michigan, http://www.sph.umich.edu/statgen/software (for SIBMED)
-
- CHLC Genetics Maps, http://lpg.nci.nih.gov/html-chlc/ChlcMaps.html
-
- Division of Statistical Genetics, Department of Human Genetics, University of Pittsburgh, http://watson.hgen.pitt.edu/mega2.html (for Mega2 version 2.2)
-
- Généthon, http://www.genethon.fr/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for AUTS1 [MIM 209850], FOXP2 [MIM 606354], and SPCH1 [MIM 602081])
References
-
- Baddeley A, Gathercole S, Papagno C (1998) The phonological loop as a language learning device. Psychol Rev 1:158–173 - PubMed
-
- Baddeley A, Wilson BA (1993) A developmental deficit in short-term phonological memory: implications for language and reading. Memory 1:65–78 - PubMed
-
- Beitchman JH, Brownlie EB, Inglis A, Wild J, Mathews R, Schachter D, Kroll R, Martin S, Ferguson B, Lancee W (1994) Seven-year follow-up of speech/language-impaired and control children: speech/language stability and outcome. J Am Acad Child Adolesc Psychiatry 33:1322–1330 - PubMed
-
- Bishop DVM, Adams C (1990) A prospective study of the relationship between specific language impairment, phonological disorders and reading retardation. J Child Psychol Psychiatry 31:1027–1050 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

