Robustness of circadian rhythms with respect to molecular noise
- PMID: 11792856
- PMCID: PMC117364
- DOI: 10.1073/pnas.022628299
Robustness of circadian rhythms with respect to molecular noise
Abstract
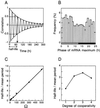
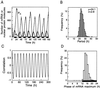
We use a core molecular model capable of generating circadian rhythms to assess the robustness of circadian oscillations with respect to molecular noise. The model is based on the negative feedback exerted by a regulatory protein on the expression of its gene. Such a negative regulatory mechanism underlies circadian oscillations of the PER protein in Drosophila and of the FRQ protein in Neurospora. The model incorporates gene transcription into mRNA, translation of mRNA into protein, reversible phosphorylation leading to degradation of the regulatory protein, transport of the latter into the nucleus, and repression of gene expression by the nuclear form of the protein. To assess the effect of molecular noise, we perform stochastic simulations after decomposing the deterministic model into elementary reaction steps. The oscillations predicted by the stochastic simulations agree with those obtained with the deterministic version of the model. We show that robust circadian oscillations can occur already with a limited number of mRNA and protein molecules, in the range of tens and hundreds, respectively. Entrainment by light/dark cycles and cooperativity in repression enhance the robustness of circadian oscillations with respect to molecular noise.
Figures



References
-
- Dunlap J C. Cell. 1999;96:271–290. - PubMed
-
- Young M W, Kay S A. Nat Rev Genet. 2001;2:702–715. - PubMed
-
- Williams J A, Sehgal A. Annu Rev Physiol. 2001;63:729–755. - PubMed
-
- Reppert S M, Weaver D R. Annu Rev Physiol. 2001;63:647–676. - PubMed
-
- Goldbeter A. Proc R Soc London Ser B. 1995;261:319–324. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

