Genomic organization and promoter analysis of the human heat shock factor 2 gene
- PMID: 11795475
- PMCID: PMC434421
- DOI: 10.1379/1466-1268(2001)006<0377:goapao>2.0.co;2
Genomic organization and promoter analysis of the human heat shock factor 2 gene
Abstract
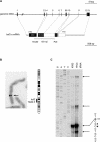
Heat shock factor 2 (HSF2) is a member of the heat shock transcription factor family, which appears to be activated during differentiation and development rather than on cellular stress. Here we report the isolation and characterization of the human hsf2 gene and its 5'-flanking region. The transcription unit of the human hsf2 gene consists of 13 exons dispersed over 33 kbp of genomic DNA on chromosome 6. The hsf2 mRNA is transcribed from multiple start sites, and initiation from the major site results in a transcript of 2.45 kb. A functional promoter, as determined by the ability to direct expression of a transiently transfected luciferase reporter gene, resides in a 950-bp upstream region of the human hsf2 gene. Examination of the core promoter sequence revealed a high GC content and lack of a canonical TATA box. This feature seems to be common among various species, as comparison of the hsf2 proximal promoter sequences from human, mouse, and rat showed distinct conserved regions. Moreover, the overall architecture of the human hsf2 gene is similar to its mouse counterpart. A comparison between human hsf2 gene and other hsf genes showed striking similarities in exon size. However, the exons are assembled in an hsf-specific manner.
Figures



References
-
- Adams MD, Celniker SE, and Holt RA. et al. 2000 The genome sequence of. Drosophila melanogaster. Science. 287:2185–2195. - PubMed
-
- Carey M, Smale ST 2000 Transcription initiation site mapping. In: Transcriptional Regulation in Eukaryotes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 116–123.
-
- Clos J, Westwood T, Becker PB, Wilson S, Lambert K, Wu C. Molecular cloning and expression of hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. - PubMed
-
- Eriksson M, Jokinen E, Sistonen L, Leppä S. Heat shock factor 2 is activated during mouse heart development. Int J Dev Biol. 2000;44:471–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
