Molecular analysis of rifampin resistance in Bacillus anthracis and Bacillus cereus
- PMID: 11796364
- PMCID: PMC127050
- DOI: 10.1128/AAC.46.2.511-513.2002
Molecular analysis of rifampin resistance in Bacillus anthracis and Bacillus cereus
Abstract
Rifampin-resistant mutants were selected from UV-light-treated Bacillus cereus (20 mutants) and attenuated B. anthracis (23 mutants). In addition, spontaneous rifampin-resistant mutants were also isolated in attenuated B. anthracis (22 mutants). The rifampin resistance clusters of the rpoB gene were sequenced for all 65 mutants. Mutations associated with resistance were consistent with those from other bacteria, though two novel changes were observed. The spontaneous rate of resistance was estimated at 1.57 x 10(-9) mutations/generation by a Luria-Delbrück fluctuation test.
Figures
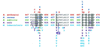
References
-
- Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA Polymerase. Cell 104:901–912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

