Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus
- PMID: 11796572
- PMCID: PMC127691
- DOI: 10.1128/IAI.70.2.470-480.2002
Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus
Abstract
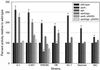
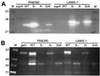
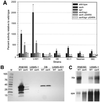
The accessory gene regulator (agr) and the staphylococcal accessory regulator (sar) are central regulatory elements that control the production of Staphylococcus aureus virulence factors. To date, the functions of these loci have been defined almost exclusively using RN6390, which is representative of the laboratory strain 8325-4. However, RN6390 was recently shown to have a mutation in rsbU that results in a phenotype resembling that of a sigB mutant (I. Kullik et al., J. Bacteriol. 180:4814-4820, 1998). For that reason, it remains unclear whether the regulatory events defined in RN6390 are representative of the events that take place in clinical isolates of S. aureus. To address this issue, we generated mutations in the sarA and agr loci of three laboratory strains (RN6390, Newman, and S6C) and four clinical isolates (UAMS-1, UAMS-601, DB, and SC-1). Mutation of sarA in the cna-positive strains UAMS-1 and UAMS-601 resulted in an increased capacity to bind collagen, while mutation of agr had little impact. Northern blot analysis confirmed that the increase in collagen binding was due to increased cna transcription. Without exception, mutation of sarA resulted in increased production of proteases and a decreased capacity to bind fibronectin. Mutation of agr had the opposite effect. Although mutation of sarA resulted in a slight reduction in fnbA transcription, changes in the ability to bind fibronectin appeared to be more directly correlated with changes in protease activity. Lipase production was reduced in both sarA and agr mutants. While mutation of sarA in RN6390 resulted in reduced hemolytic activity, it had the opposite effect in all other strains. There appeared to be reduced levels of the sarC transcript in RN6390, but there was no difference in the overall pattern of sar transcription or the production of SarA. Although mutation of sarA resulted in decreased RNAIII transcription, this effect was not evident under all growth conditions. Taken together, these results suggest that studies defining the regulatory roles of sarA and agr by using RN6390 are not always representative of the events that occur in clinical isolates of S. aureus.
Figures



References
-
- Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317–326. - PubMed
-
- Bohach, G. A., and T. J. Foster. 2000. Staphylococcus aureus exotoxins, p.367–378. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

