Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease
- PMID: 11796592
- PMCID: PMC127674
- DOI: 10.1128/IAI.70.2.631-641.2002
Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease
Abstract
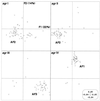
The expression of most Staphylococcus aureus virulence factors is controlled by the agr locus, which encodes a two-component signaling pathway whose activating ligand is an agr-encoded autoinducing peptide (AIP). A polymorphism in the amino acid sequence of the AIP and of its corresponding receptor divides S. aureus strains into four major groups. Within a given group, each strain produces a peptide that can activate the agr response in the other member strains, whereas the AIPs belonging to different groups are usually mutually inhibitory. We investigated a possible relationship between agr groups and human S. aureus disease by studying 198 S. aureus strains isolated from 14 asymptomatic carriers, 66 patients with suppurative infection, and 114 patients with acute toxemia. The agr group and the distribution of 24 toxin genes were analyzed by PCR, and the genetic background was determined by means of amplified fragment length polymorphism (AFLP) analysis. The isolates were relatively evenly distributed among the four agrgroups, with 61 strains belonging to agr group I, 49 belonging to group II, 43 belonging to group III, and 45 belonging to group IV. Principal coordinate analysis performed on the AFLP distance matrix divided the 198 strains into three main phylogenetic groups, AF1 corresponding to strains of agr group IV, AF2 corresponding to strains of agr groups I and II, and AF3 corresponding to strains of agr group III. This indicated that the agr type was linked to the genetic background. A relationship between genetic background, agr group, and disease type was observed for several toxin-mediated diseases: for instance, agr group IV strains were associated with generalized exfoliative syndromes, and phylogenetic group AF1 strains with bullous impetigo. Among the suppurative infections, endocarditis strains mainly belonged to phylogenetic group AF2 and agr groups I and II. While these results do not show a direct role of the agr type in the type of human disease caused by S. aureus, the agr group may reflect an ancient evolutionary division of S. aureus in terms of this species' fundamental biology.
Figures





References
-
- Arbuthnott, J. P., D. C. Coleman, and J. S. de Azavedo. 1990. Staphylococcal toxins in human disease. Soc. Appl. Bacteriol. Symp. Ser. 19:101S–107S. - PubMed
-
- Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642–650. - PubMed
-
- Day, N. P., C. E. Moore, M. C. Enright, A. R. Berendt, J. M. Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114–116. - PubMed
-
- De Buyser, M. L., A. Morvan, F. Grimont, and N. El Solh. 1989. Characterization of Staphylococcus species by ribosomal RNA gene restriction patterns. J. Gen. Microbiol. 135:989–999. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

