The mitotic spindle is required for loading of the DASH complex onto the kinetochore
- PMID: 11799062
- PMCID: PMC155319
- DOI: 10.1101/gad.959402
The mitotic spindle is required for loading of the DASH complex onto the kinetochore
Abstract
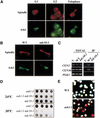
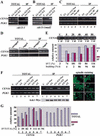
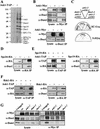
A role for the mitotic spindle in the maturation of the kinetochore has not been defined previously. Here we describe the isolation of a novel and conserved essential gene, ASK1, from Saccharomyces cerevisiae involved in this process. ask1 mutants display either G(2)/M arrest or segregation of DNA masses without the separation of sister chromatids, resulting in massive nondisjunction and broken spindles. Ask1 localizes along mitotic spindles and to kinetochores, and cross-links to centromeric DNA. Microtubules are required for Ask1 binding to kinetochores, and are partially required to maintain its association. We found Ask1 is part of a multisubunit complex, DASH, that contains approximately 10 components, including several proteins essential for mitosis including Dam1, Duo1, Spc34, Spc19, and Hsk1. The Ipl1 kinase controls the phosphorylation of Dam1 in the DASH complex and may regulate its function. We propose that DASH is a microtubule-binding complex that is transferred to the kinetochore prior to mitosis, thereby defining a new step in kinetochore maturation.
Figures

References
-
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJH, Bousset K, Kanji Furuya K, Diffley JFX, Carr A, Elledge SJ. Mrc1 transduces DNA replication stress signals to activate Rad53. Nat Cell Biol. 2001;3:958–965. - PubMed
-
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
