E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints
- PMID: 11799067
- PMCID: PMC155321
- DOI: 10.1101/gad.949802
E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints
Abstract
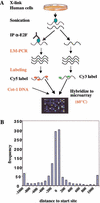
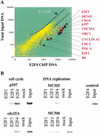
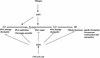
The E2F transcription factor family is known to play a key role in the timely expression of genes required for cell cycle progression and proliferation, but only a few E2F target genes have been identified. We explored the possibility that E2F regulators play a broader role by identifying additional genes bound by E2F in living human cells. A protocol was developed to identify genomic binding sites for DNA-binding factors in mammalian cells that combines immunoprecipitation of cross-linked protein-DNA complexes with DNA microarray analysis. Among approximately 1200 genes expressed during cell cycle entry, we found that the promoters of 127 were bound by the E2F4 transcription factor in primary fibroblasts. A subset of these targets was also bound by E2F1. Most previously identified target genes known to have roles in DNA replication and cell cycle control and represented on the microarray were confirmed by this analysis. We also identified a remarkable cadre of genes with no previous connection to E2F regulation, including genes that encode components of the DNA damage checkpoint and repair pathways, as well as factors involved in chromatin assembly/condensation, chromosome segregation, and the mitotic spindle checkpoint. Our data indicate that E2F directly links cell cycle progression with the coordinate regulation of genes essential for both the synthesis of DNA as well as its surveillance.
Figures


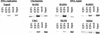

References
-
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. - PubMed
-
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. - PubMed
-
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
