The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation
- PMID: 11799113
- PMCID: PMC1424220
- DOI: 10.1074/jbc.M108740200
The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation
Abstract
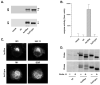
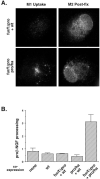
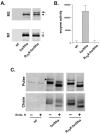
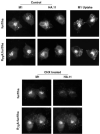
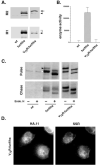
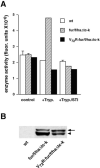
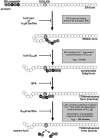
The propeptide of furin has multiple roles in guiding the activation of the endoprotease in vivo. The 83-residue N-terminal propeptide is autoproteolytically excised in the endoplasmic reticulum (ER) at the consensus furin site, -Arg(104)-Thr-Lys-Arg(107)-, but remains bound to furin as a potent autoinhibitor. Furin lacking the propeptide is ER-retained and proteolytically inactive. Co-expression with the propeptide, however, restores trans-Golgi network (TGN) localization and enzyme activity, indicating that the furin propeptide is an intramolecular chaperone. Blocking this step results in localization to the ER-Golgi intermediate compartment (ERGIC)/cis-Golgi network (CGN), suggesting the ER and ERGIC/CGN recognize distinct furin folding intermediates. Following transport to the acidified TGN/endosomal compartments, furin cleaves the bound propeptide at a second, internal P1/P6 Arg site (-Arg-Gly-Val(72)-Thr-Lys-Arg(75)-) resulting in propeptide dissociation and enzyme activation. Cleavage at Arg(75), however, is not required for proper furin trafficking. Kinetic analyses of peptide substrates indicate that the sequential pH-modulated propeptide cleavages result from the differential recognition of these sites by furin. Altering this preference by converting the internal site to a canonical P1/P4 Arg motif (Val(72) --> Arg) caused ER retention and blocked activation of furin, demonstrating that the structure of the furin propeptide mediates folding of the enzyme and directs its pH-regulated, compartment-specific activation in vivo.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

