Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity
- PMID: 11799165
- PMCID: PMC135892
- DOI: 10.1128/jvi.76.4.1697-1706.2002
Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity
Abstract
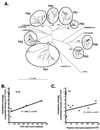
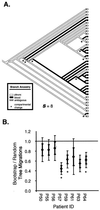
We have recently reported an increased heterogeneity in the human immunodeficiency virus type 1 (HIV-1) envelope gene (env) in HIV-1-infected patients with pulmonary tuberculosis (TB) compared to patients with HIV-1 alone. This increase may be a result of dissemination of lung-derived HIV-1 isolates from sites of Mycobacterium tuberculosis infection and/or the systemic activation of the immune system in response to TB. To distinguish between these two mechanisms, blood and pleural fluid samples were obtained from HIV-1-infected patients with active pleural TB in Kampala, Uganda (CD4 cell counts of 34 to 705 cells/microl, HIV-1 plasma loads of 2,400 to 280,000 RNA copies/ml, and HIV-1 pleural loads of 7,600 to 4,500,000 RNA copies/ml). The C2-C3 coding region of HIV-1 env was PCR amplified from lysed peripheral blood mononuclear cells and pleural fluid mononuclear cells and reverse transcriptase-PCR amplified from plasma and pleural fluid HIV-1 virions of eight HIV-1 patients with pleural TB. Phylogenetic and phenetic analyses revealed a compartmentalization of HIV-1 quasispecies between blood and pleural space in four of eight patients, with migration events between the compartments. There was a trend for a greater genetic heterogeneity in the pleural space, which may be the result of an M. tuberculosis-mediated increase in HIV-1 replication and/or selection pressure at the site of infection. Collectively, these findings suggest that HIV-1 quasispecies in the M. tuberculosis-infected pleural space may leak into the systemic circulation and lead to increased systemic HIV-1 heterogeneity during TB.
Figures



References
-
- Ait-Khaled, M., J. E. McLaughlin, M. A. Johnson, and V. C. Emery. 1995. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS 9:675-683. - PubMed
-
- Antony, V. B., S. W. Godbey, S. L. Kunkel, J. W. Hott, D. L. Hartman, M. D. Burdick, and R. M. Strieter. 1993. Recruitment of inflammatory cells to the pleural space. Chemotactic cytokines, IL-8, and monocyte chemotactic peptide-1 in human pleural fluids. J. Immunol. 151:7216-7223. - PubMed
-
- Beck, J. M. 1998. Pleural disease in patients with acquired immune deficiency syndrome. Clin. Chest Med. 19:341-349. - PubMed
-
- Boyum, A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Isolation of mononuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. Suppl. 97:77-89. - PubMed
-
- Collins, K. R., H. Mayanja-Kizza, B. A. Sullivan, M. E. Quinones-Mateu, Z. Toossi, and E. J. Arts. 2000. Greater diversity of HIV-1 quasispecies in HIV-infected individuals with active tuberculosis. J. Acquir. Immune Defic. Syndr. 24:408-417. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

