Construction and characterization of a replication-competent retroviral shuttle vector plasmid
- PMID: 11799171
- PMCID: PMC135915
- DOI: 10.1128/jvi.76.4.1762-1768.2002
Construction and characterization of a replication-competent retroviral shuttle vector plasmid
Abstract
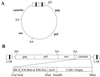
We constructed two versions of an RCASBP-based retroviral shuttle vector, RSVP (RCASBP shuttle vector plasmid), containing either the zeocin or blasticidin resistance gene. In this vector, the drug resistance gene is expressed in avian cells from the long terminal repeat (LTR) promoter, whereas in bacteria the resistance gene is expressed from a bacterial promoter. The vector contains a bacterial origin of replication (ColE1) to allow circular viral DNA to replicate as a plasmid in bacteria. The vector also contains the lac operator sequence, which binds to the lac repressor protein, providing a simple and rapid way to purify the vector DNA. The RSVP plasmid contains the following sequence starting with the 5" end: LTR, gag, pol, env, drug resistance gene, lac operator, ColE1, LTR. After this plasmid was transfected into DF-1 cells, we were able to rescue the circularized unintegrated viral DNA from RSVP simply by transforming the Hirt DNA into Escherichia coli. Furthermore, we were able to rescue the integrated provirus. DNA from infected cells was digested with an appropriate restriction enzyme (ClaI) and the vector-containing segments were enriched using lac repressor protein and then self-ligated. These enriched fractions were used to transform E. coli. The transformation was successful and we did recover integration sites, but higher-efficiency rescue was obtained with electroporation. The vector is relatively stable upon passage in avian cells. Southern blot analyses of genomic DNAs derived from successive viral passages under nonselective conditions showed that the cassette (drug resistance gene-lac operator-ColE1) insert was present in the vector up to the third viral passage for both resistance genes, which suggests that the RSVP vectors are stable for approximately three viral passages. Together, these results showed that RSVP vectors are useful tools for cloning unintegrated or integrated viral DNAs.
Figures





Similar articles
-
Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants.J Virol. 1990 Nov;64(11):5475-84. doi: 10.1128/JVI.64.11.5475-5484.1990. J Virol. 1990. PMID: 2170682 Free PMC article.
-
Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors.Mol Cell Biol. 1983 Dec;3(12):2241-9. doi: 10.1128/mcb.3.12.2241-2249.1983. Mol Cell Biol. 1983. PMID: 6318091 Free PMC article.
-
Molecular cloning of the Mason-Pfizer monkey virus genome: characterization and cloning of subgenomic fragments.Virology. 1985 Apr 30;142(2):223-40. doi: 10.1016/0042-6822(85)90331-9. Virology. 1985. PMID: 2997984
-
Vaccinia viral/retroviral chimeric vectors.Curr Gene Ther. 2004 Dec;4(4):417-26. doi: 10.2174/1566523043346101. Curr Gene Ther. 2004. PMID: 15578991 Review.
-
Shuttle vectors for studying mutagenesis in mammalian cells.J Photochem Photobiol B. 1989 Apr;3(2):143-55. doi: 10.1016/1011-1344(89)80057-0. J Photochem Photobiol B. 1989. PMID: 2542504 Review.
Cited by
-
Rous sarcoma virus (RSV) integration in vivo: a CA dinucleotide is not required in U3, and RSV linear DNA does not autointegrate.J Virol. 2008 Jan;82(1):503-12. doi: 10.1128/JVI.01441-07. Epub 2007 Oct 24. J Virol. 2008. PMID: 17959663 Free PMC article.
-
Mutations in the U5 region adjacent to the primer binding site affect tRNA cleavage by human immunodeficiency virus type 1 reverse transcriptase in vivo.J Virol. 2008 Jan;82(2):719-27. doi: 10.1128/JVI.02611-06. Epub 2007 Nov 7. J Virol. 2008. PMID: 17989171 Free PMC article.
-
Addition of N-terminal peptide sequences activates the oncogenic and signaling potentials of the catalytic subunit p110α of phosphoinositide-3-kinase.Cell Cycle. 2011 Nov 1;10(21):3731-9. doi: 10.4161/cc.10.21.17920. Epub 2011 Nov 1. Cell Cycle. 2011. PMID: 22045127 Free PMC article.
-
Alternate polypurine tracts affect rous sarcoma virus integration in vivo.J Virol. 2006 Oct;80(20):10281-4. doi: 10.1128/JVI.00361-06. J Virol. 2006. PMID: 17005708 Free PMC article.
-
Alternate polypurine tracts (PPTs) affect the rous sarcoma virus RNase H cleavage specificity and reveal a preferential cleavage following a GA dinucleotide sequence at the PPT-U3 junction.J Virol. 2005 Nov;79(21):13694-704. doi: 10.1128/JVI.79.21.13694-13704.2005. J Virol. 2005. PMID: 16227289 Free PMC article.
References
-
- Berdy, J. 1980. Handbook of antibiotic compounds, vol. 4, p. 459-491. CRC Press, Boca Raton, Fla.
-
- Calmels, T., M. Parriche, H. Burand, and G. Tiraby. 1991. High efficiency transformation of Tolypocladium geodes conidiospores to phleomycin resistance. Curr. Genet. 20:309-314. - PubMed
-
- Gatignol, A., M. Baron, and G. Tiraby. 1987. Pleomycin resistance encoded by the ble gene from transposon Tn5 as a dominant selectable marker in Saccharomyces cerevisiae. Mol. Gen. Genet. 207:342-348. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

