The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection
- PMID: 11799183
- PMCID: PMC135906
- DOI: 10.1128/jvi.76.4.1884-1891.2002
The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection
Abstract
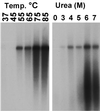
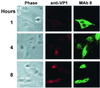
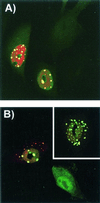
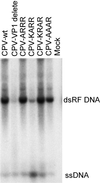
The unique N-terminal region of the parvovirus VP1 capsid protein is required for infectivity by the capsids but is not required for capsid assembly. The VP1 N terminus contains a number of groups of basic amino acids which resemble classical nuclear localization sequences, including a conserved sequence near the N terminus comprised of four basic amino acids, which in a peptide can act to transport other proteins into the cell nucleus. Testing with a monoclonal antibody recognizing residues 2 to 13 of VP1 (anti-VP1-2-13) and with a rabbit polyclonal serum against the entire VP1 unique region showed that the VP1 unique region was not exposed on purified capsids but that it became exposed after treatment of the capsids with heat (55 to 75 degrees C), or urea (3 to 5 M). A high concentration of anti-VP1-2-13 neutralized canine parvovirus (CPV) when it was incubated with the virus prior to inoculation of cells. Both antibodies blocked infection when injected into cells prior to virus inoculation, but neither prevented infection by coinjected infectious plasmid DNA. The VP1 unique region could be detected 4 and 8 h after the virus capsids were injected into cells, and that sequence exposure appeared to be correlated with nuclear transport of the capsids. To examine the role of the VP1 N terminus in infection, we altered that sequence in CPV, and some of those changes made the capsids inefficient at cell infection.
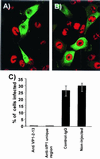
Figures
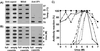
Similar articles
-
Assaying for structural variation in the parvovirus capsid and its role in infection.Virology. 1998 Oct 10;250(1):106-17. doi: 10.1006/viro.1998.9352. Virology. 1998. PMID: 9770425
-
Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport.J Virol. 2000 May;74(10):4853-9. doi: 10.1128/jvi.74.10.4853-4859.2000. J Virol. 2000. PMID: 10775624 Free PMC article.
-
Canine parvovirus capsid assembly and differences in mammalian and insect cells.Virology. 2001 Jan 20;279(2):546-57. doi: 10.1006/viro.2000.0734. Virology. 2001. PMID: 11162810
-
The role of nuclear localization signal in parvovirus life cycle.Virol J. 2017 Apr 14;14(1):80. doi: 10.1186/s12985-017-0745-1. Virol J. 2017. PMID: 28410597 Free PMC article. Review.
-
Role of capsid proteins in parvoviruses infection.Virol J. 2015 Aug 4;12:114. doi: 10.1186/s12985-015-0344-y. Virol J. 2015. PMID: 26239432 Free PMC article. Review.
Cited by
-
Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges.J Virol. 2006 Sep;80(17):8482-92. doi: 10.1128/JVI.00683-06. J Virol. 2006. PMID: 16912298 Free PMC article.
-
The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor.J Virol. 2003 Feb;77(3):1718-26. doi: 10.1128/jvi.77.3.1718-1726.2003. J Virol. 2003. PMID: 12525605 Free PMC article.
-
Visualization of the externalized VP2 N termini of infectious human parvovirus B19.J Virol. 2008 Aug;82(15):7306-12. doi: 10.1128/JVI.00512-08. Epub 2008 May 28. J Virol. 2008. PMID: 18508892 Free PMC article.
-
Detecting small changes and additional peptides in the canine parvovirus capsid structure.J Virol. 2008 Nov;82(21):10397-407. doi: 10.1128/JVI.00972-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701590 Free PMC article.
-
Parvovirus particles and movement in the cellular cytoplasm and effects of the cytoskeleton.Virology. 2014 May;456-457:342-52. doi: 10.1016/j.virol.2014.04.003. Epub 2014 Apr 25. Virology. 2014. PMID: 24889253 Free PMC article.
References
-
- Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. - PubMed
-
- Anderson, S., M. Momoeda, M. Kawase, S. Kajigaya, and N. S. Young. 1995. Peptides derived from the unique region of B19 parvovirus minor capsid protein elicit neutralizing antibodies in rabbits. Virology 206:626-632. - PubMed
-
- Casal, J. I., J. P. Langeveld, E. Cortes, W. W. Schaaper, E. van Dijk, C. Vela, S. Kamstrup, and R. H. Meloen. 1995. Peptide vaccine against canine parvovirus: identification of two neutralization subsites in the N terminus of VP2 and optimization of the amino acid sequence. J. Virol. 69:7274-7277. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

